3CC0
 
 | |
6LL9
 
 | |
2ZEU
 
 | | S. Cerevisiae Geranylgeranyl Pyrophosphate Synthase in Complex with BPH-715 | | Descriptor: | 3-(DECYLOXY)-1-(2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Geranylgeranyl pyrophosphate synthetase | | Authors: | Guo, R.T, Chen, C.K.-M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation
J.Am.Chem.Soc., 131, 2009
|
|
2ZEV
 
 | | S. Cerevisiae Geranylgeranyl Pyrophosphate Synthase in Complex with Magnesium, IPP and BPH-715 | | Descriptor: | 3-(DECYLOXY)-1-(2,2-DIPHOSPHONOETHYL)PYRIDINIUM, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, ... | | Authors: | Guo, R.T, Chen, C.K.-M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation
J.Am.Chem.Soc., 131, 2009
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLK
 
 | |
5VLP
 
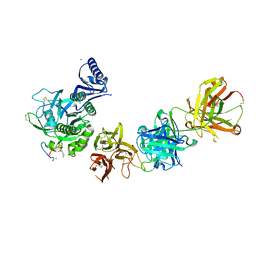 | | PCSK9 complex with LDLR antagonist peptide and Fab7G7 | | Descriptor: | Fab7G7 heavy chain, Fab7G7 light chain, LDLR antagonist peptide, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLA
 
 | | Short PCSK9 delta-P' complex with Fusion2 peptide | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-MET-PRO-TRP-ASN-LEU-VAL-ARG-ILE-GLY-LEU-LEU | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7VWP
 
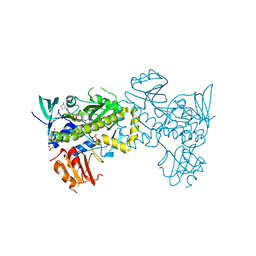 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
5VLH
 
 | | Short PCSK9 delta-P' complex with peptide Pep1 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-ARG-LEU-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-PRO-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLL
 
 | | Short PCSK9 delta-P' complex with peptide Pep3 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-PHE-ILE-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-LEU-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6A9Y
 
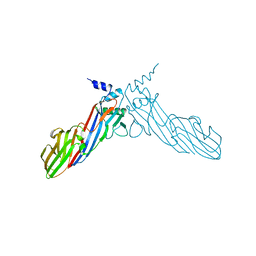 | | The crystal structure of Mu homology domain of SGIP1 | | Descriptor: | SH3-containing GRB2-like protein 3-interacting protein 1 | | Authors: | Feng, Y, Liu, X. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SGIP1 dimerizes via intermolecular disulfide bond in mu HD domain during cellular endocytosis.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6J2R
 
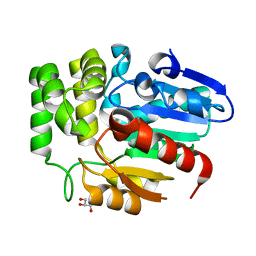 | | Crystal structure of Striga hermonthica HTL8 (ShHTL8) | | Descriptor: | GLYCEROL, Hyposensitive to light 8 | | Authors: | Zhang, Y.Y, Xi, Z. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6VPT
 
 | |
6GFM
 
 | | Crystal structure of the Escherichia coli nucleosidase PpnN (pppGpp-form) | | Descriptor: | Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Zhang, Y, Baerentsen, R.L, Gerdes, K, Brodersen, D.E. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch.
Mol.Cell, 74, 2019
|
|
6GFL
 
 | |
4HK2
 
 | | U7Ub25.2540 | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
4HJK
 
 | | U7Ub7 Disulfide variant | | Descriptor: | PHOSPHATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
4NMX
 
 | |
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
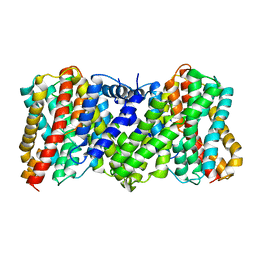 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8H97
 
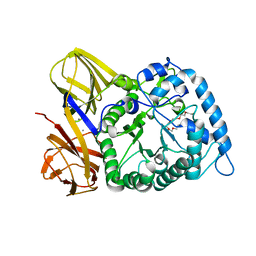 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
5TIG
 
 | |
6BGN
 
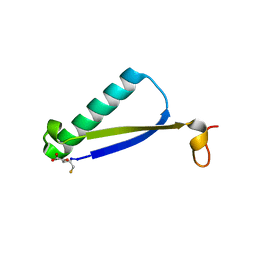 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
6JJI
 
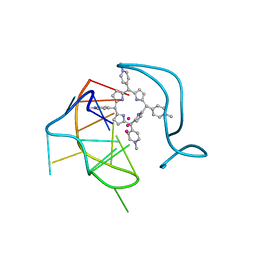 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
