9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
5KF4
 
 | |
6CK8
 
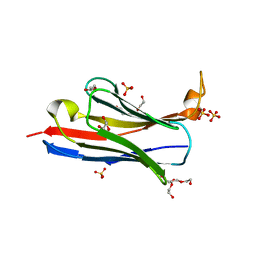 | |
5E2V
 
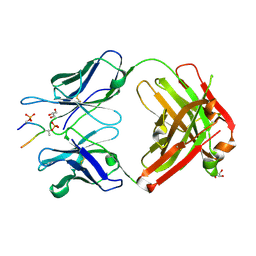 | |
5E2W
 
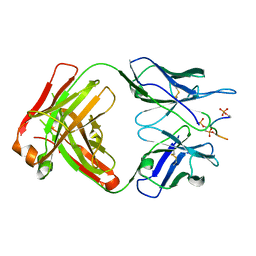 | | Anti-TAU AT8 FAB with triply phosphorylated TAU peptide | | Descriptor: | AT8 HEAVY CHAIN, AT8 LIGHT CHAIN, TAU-PHOSPHOPEPTIDE | | Authors: | Malia, T, Teplyakov, A. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Proteins, 84, 2016
|
|
5E2U
 
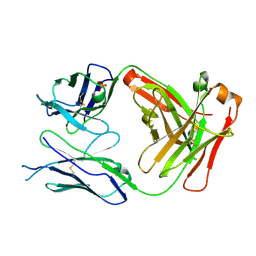 | |
5E2T
 
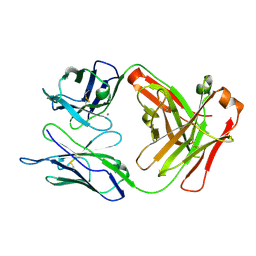 | | Crystal structure of anti-TAU antibody AT8 FAB | | Descriptor: | AT8 HEAVY CHAIN, AT8 LIGHT CHAIN, CALCIUM ION | | Authors: | Malia, T, Teplyakov, A. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Proteins, 84, 2016
|
|
8F43
 
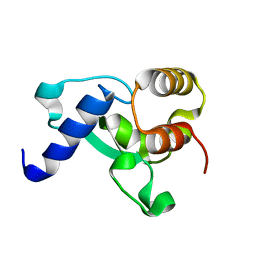 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8JNI
 
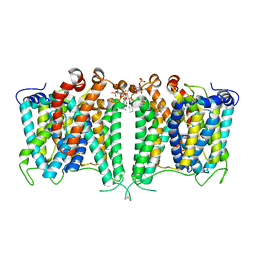 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8JNJ
 
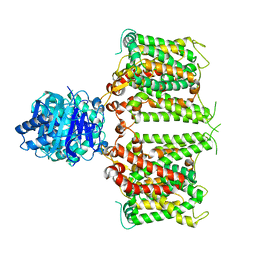 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7YQE
 
 | |
5FDP
 
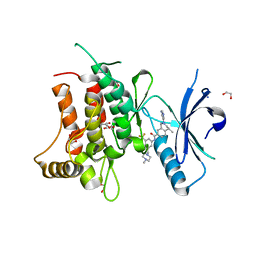 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
7YT9
 
 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
8WZB
 
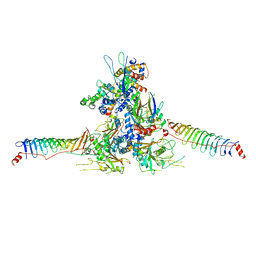 | | RS head-neck monomer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
8X2U
 
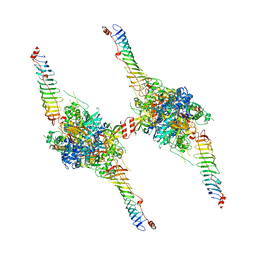 | | Radial spoke head-neck dimer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
3MCK
 
 | |
5WXD
 
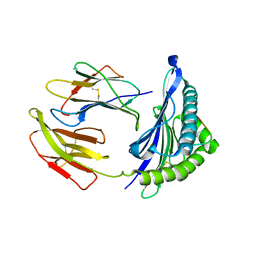 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, H7-25, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5ZWZ
 
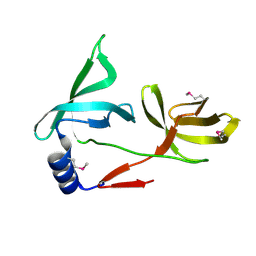 | |
5ZWX
 
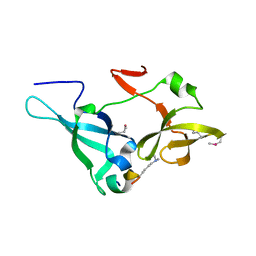 | |
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
