3OMG
 
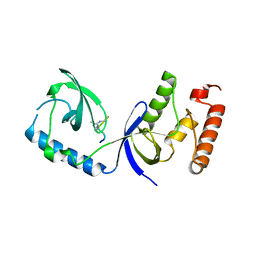 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ULP
 
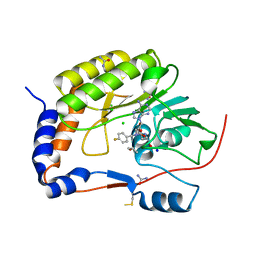 | | Structure of the NS5 methyltransferase from Zika bound to MS2042 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][(4-fluorophenyl)methyl]amino}-5'-deoxyadenosine, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Jain, R, Aggarwal, A.K. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a S-adenosylmethionine analog that intrudes the RNA-cap binding site of Zika methyltransferase.
Sci Rep, 7, 2017
|
|
5CZK
 
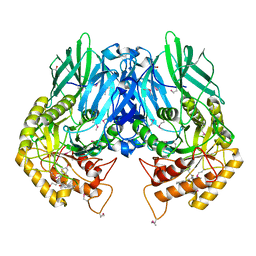 | | Structure of E. coli beta-glucuronidase bound with a novel, potent inhibitor 1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea | | Descriptor: | 1-[(6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Roberts, A.R, Wallace, B.R, Redinbo, M.R. | | Deposit date: | 2015-07-31 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity.
Chem.Biol., 22, 2015
|
|
5D8W
 
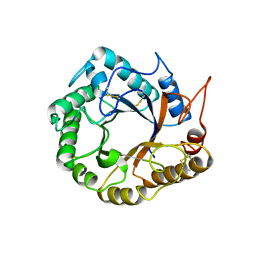 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5D8Z
 
 | | Structrue of a lucidum protein | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5WYZ
 
 | | Crystal structure of human TLR8 in complex with CU-CPT9b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-methyl-4-oxidanyl-phenyl)quinolin-7-ol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
5V2N
 
 | | Crystal Structure of APO Human SETD8 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase KMT5A | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
5WYX
 
 | | Crystal structure of human TLR8 in complex with CU-CPT8m | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(3-methylphenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
4MFI
 
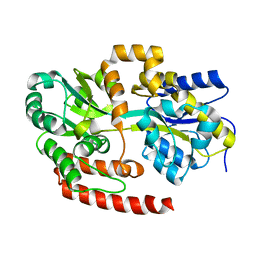 | | Crystal structure of Mycobacterium tuberculosis UgpB | | Descriptor: | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UspB | | Authors: | Jiang, D, Bartlam, M, Rao, Z. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Febs J., 281, 2014
|
|
7R1U
 
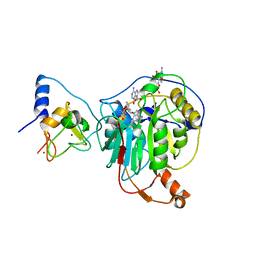 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
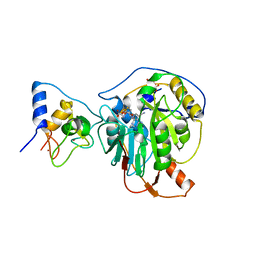 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
4LG6
 
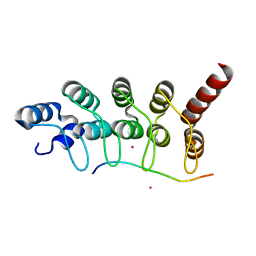 | | Crystal structure of ANKRA2-CCDC8 complex | | Descriptor: | Ankyrin repeat family A protein 2, Coiled-coil domain-containing protein 8, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Bian, C, Tempel, W, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ankyrin Repeats of ANKRA2 Recognize a PxLPxL Motif on the 3M Syndrome Protein CCDC8.
Structure, 23, 2015
|
|
4FL6
 
 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
6BJQ
 
 | |
6D4O
 
 | | Eubacterium eligens beta-glucuronidase bound to an amoxapine-glucuronide conjugate | | Descriptor: | (5aR,9aR)-2-chloro-11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5a,6,9,9a-tetrahydrodibenzo[b,f][1,4]oxazepine, Beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Pellock, S.J, Walton, W.G, Redinbo, M.R. | | Deposit date: | 2018-04-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut Microbial beta-Glucuronidase Inhibition via Catalytic Cycle Interception.
ACS Cent Sci, 4, 2018
|
|
6BJW
 
 | |
6BO6
 
 | | Eubacterium eligens beta-glucuronidase bound to UNC4917 glucuronic acid conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycoside Hydrolase Family 2 candidate b-glucuronidase | | Authors: | Pellock, S.J, Walton, W.G, Redinbo, M.R. | | Deposit date: | 2017-11-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Gut Microbial beta-Glucuronidase Inhibition via Catalytic Cycle Interception.
ACS Cent Sci, 4, 2018
|
|
4JKL
 
 | |
4JKM
 
 | |
4JKK
 
 | |
4IL1
 
 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
6I8L
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
5I78
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
