6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
3PP4
 
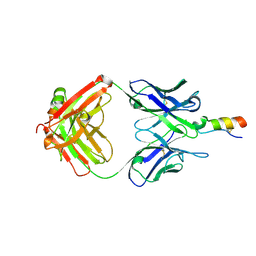 | |
3PP3
 
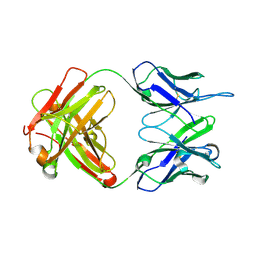 | |
7MHU
 
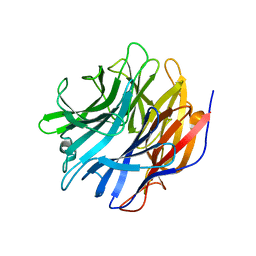 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6TW2
 
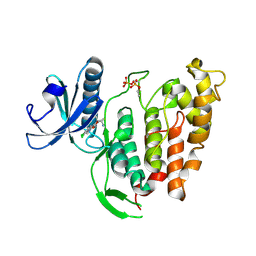 | | Re-refined crystal structure of di-phosphorylated human CLK1 in complex with a novel substituted indole inhibitor | | Descriptor: | Dual specificity protein kinase CLK1, ethyl 3-[(E)-2-amino-1-cyanoethenyl]-6,7-dichloro-1-methyl-1H-indole-2-carboxylate | | Authors: | Loll, B, Haltenhof, T, Heyd, F, Wahl, M.C. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression.
Mol.Cell, 78, 2020
|
|
1I9F
 
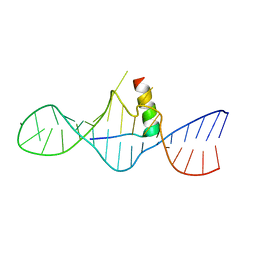 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
7BW2
 
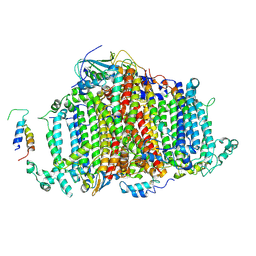 | | Crystal Structure of Cyanobacterial PSI Monomer from T.elongatus at 6.5 A Resolution | | Descriptor: | Photosystem I 4.8K protein, Photosystem I P700 chlorophyll a apoprotein A1, Photosystem I P700 chlorophyll a apoprotein A2, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Eithar, E.M, Mian, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
4HC4
 
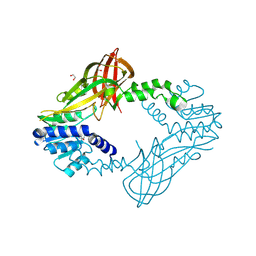 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4Q3M
 
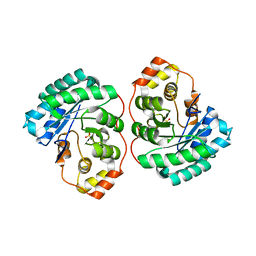 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3K
 
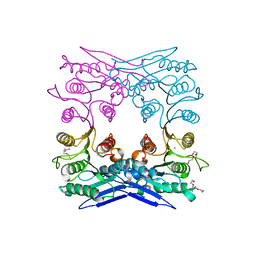 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
8RC0
 
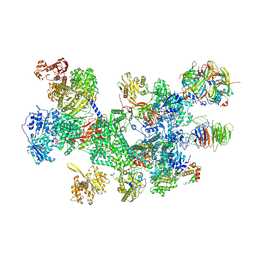 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6Q7M
 
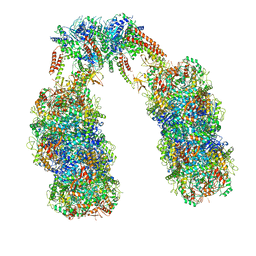 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
6Q7L
 
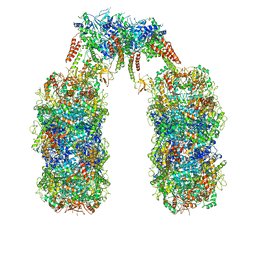 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
3FPU
 
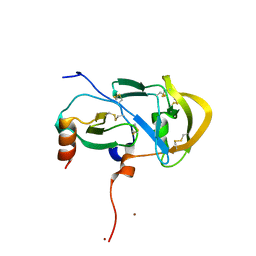 | |
3FPR
 
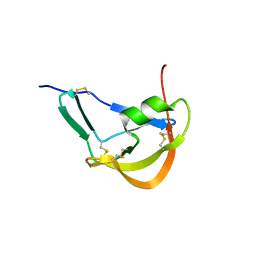 | | Crystal Structure of Evasin-1 | | Descriptor: | Evasin-1 | | Authors: | Dias, J.M, Shaw, J.P. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
6Y3X
 
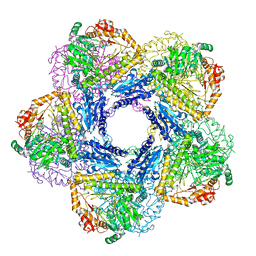 | |
3FPT
 
 | | The Crystal Structure of the Complex between Evasin-1 and CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Evasin-1 | | Authors: | Shaw, J.P, Dias, J.M. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
5DZL
 
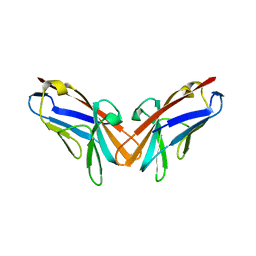 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
3JSB
 
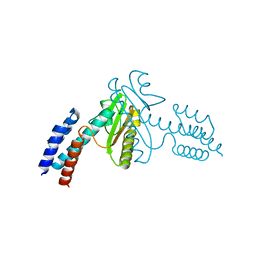 | | Crystal structure of the N-terminal domain of the Lymphocytic Choriomeningitis Virus L protein | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Morin, B, Jamal, S, Ferron, F.P, Coutard, B, Bricogne, G, Canard, B, Vonrhein, C, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription
Plos Pathog., 6, 2010
|
|
4EQV
 
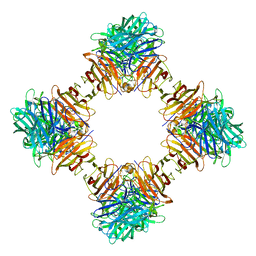 | |
4N3W
 
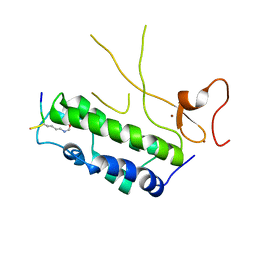 | |
4N4F
 
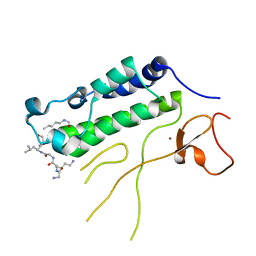 | |
2FAV
 
 | |
