7JTG
 
 | |
3U6B
 
 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
3U6K
 
 | | Ef-tu (escherichia coli) in complex with nvp-ldk733 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
2XXN
 
 | | Structure of the vIRF4-HAUSP TRAF domain complex | | Descriptor: | K10, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 7 | | Authors: | Choi, W.C, Hwang, J, Kim, M.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bilateral Inhibition of Hausp Deubiquitinase by a Viral Interferon Regulatory Factor Protein
Nat.Struct.Mol.Biol., 18, 2011
|
|
3UK8
 
 | |
3UGC
 
 | | Structural basis of Jak2 inhibition by the type II inhibtor NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Scheufler, C, Tavares, G.A, Manley, P.W, Pissot-Soldermann, C, Kroemer, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Cancer Discov, 2, 2012
|
|
3TXO
 
 | | PKC eta kinase in complex with a naphthyridine | | Descriptor: | 2-methyl-N~1~-[3-(pyridin-4-yl)-2,6-naphthyridin-1-yl]propane-1,2-diamine, Protein kinase C eta type | | Authors: | Stark, W, Rummel, G, Cowan-Jacob, S.W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4GFD
 
 | | Thymidylate kinase (TMK) from S. Aureus in complex with TK-666 | | Descriptor: | 2-(3-bromophenoxy)-4-{(1R)-3,3-dimethyl-1-[(3S)-3-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)piperidin-1-yl]butyl}benzoic acid, Thymidylate kinase | | Authors: | Olivier, N.B, Martinez-Botella, G, Keating, T. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Vivo Validation of Thymidylate Kinase (TMK) with a Rationally Designed, Selective Antibacterial Compound.
Acs Chem.Biol., 7, 2012
|
|
3S88
 
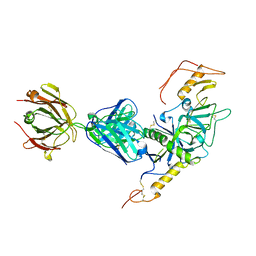 | | Crystal structure of Sudan Ebolavirus Glycoprotein (strain Gulu) bound to 16F6 | | Descriptor: | 16F6 - Heavy chain, 16F6 - Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Dias, J.M, Bale, S. | | Deposit date: | 2011-05-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.351 Å) | | Cite: | A shared structural solution for neutralizing ebolaviruses.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HLS
 
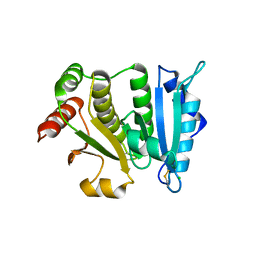 | |
2KSE
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A4C
 
 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
2MJ6
 
 | |
2M36
 
 | |
2Y34
 
 | |
2Y33
 
 | |
3DHJ
 
 | | Beta 2 microglobulin mutant W60C | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
4MWF
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2013-09-24 | | Release date: | 2013-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Hepatitis C virus e2 envelope glycoprotein core structure.
Science, 342, 2013
|
|
5OER
 
 | | Hen egg-white lysozyme refined against 5000 9 keV diffraction patterns | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C, ... | | Authors: | Gorel, A, Schlichting, I. | | Deposit date: | 2017-07-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multi-wavelength anomalous diffraction de novo phasing using a two-colour X-ray free-electron laser with wide tunability.
Nat Commun, 8, 2017
|
|
5O07
 
 | |
4N0Y
 
 | |
2V6I
 
 | | Kokobera Virus Helicase | | Descriptor: | PYROPHOSPHATE 2-, RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
3DHM
 
 | | Beta 2 microglobulin mutant D59P | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
4PX6
 
 | | SYK catalytic domain in complex with a potent pyridopyrimidinone inhibitor | | Descriptor: | 7-{[(1R,2S)-2-aminocyclohexyl]amino}-5-(1H-indol-7-ylamino)pyrido[4,3-d]pyrimidin-4(3H)-one, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Syk inhibitors with high potency in presence of blood.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2V6J
 
 | | Kokobera Virus Helicase: Mutant Met47Thr | | Descriptor: | RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
