6ANO
 
 | |
1MWP
 
 | | N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN | | Descriptor: | AMYLOID A4 PROTEIN | | Authors: | Rossjohn, J, Cappai, R, Feil, S.C, Henry, A, McKinstry, W.J, Galatis, D, Hesse, L, Multhaup, G, Beyreuther, K, Masters, C.L, Parker, M.W. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein.
Nat.Struct.Biol., 6, 1999
|
|
1MXF
 
 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
6AWI
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-N5Pan | | Descriptor: | (2R)-2-hydroxy-3,3-dimethyl-N-[3-oxo-3-(pentylamino)propyl]butanamide, ADENOSINE-5'-DIPHOSPHATE, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
1CW3
 
 | | HUMAN MITOCHONDRIAL ALDEHYDE DEHYDROGENASE COMPLEXED WITH NAD+ AND MN2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, MITOCHONDRIAL ALDEHYDE DEHYDROGENASE, ... | | Authors: | Ni, L, Zhou, J, Hurley, T.D, Weiner, H. | | Deposit date: | 1999-08-25 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms.
Protein Sci., 8, 1999
|
|
4YXX
 
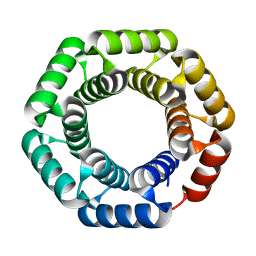 | |
4Z2B
 
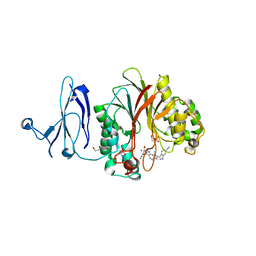 | | The structure of human PDE12 residues 161-609 in complex with GSK3036342A | | Descriptor: | 1,2-ETHANEDIOL, 2',5'-phosphodiesterase 12, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
1D1P
 
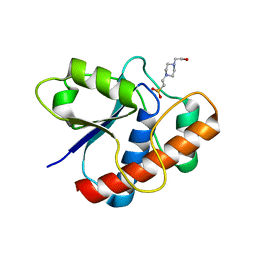 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1N2V
 
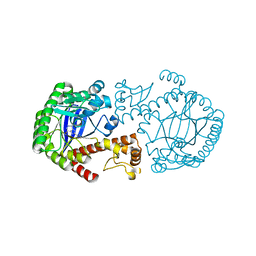 | | Crystal Structure of TGT in complex with 2-Butyl-5,6-dihydro-1H-imidazo[4,5-d]pyridazine-4,7-dione | | Descriptor: | 2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Naerum, L, Graedler, U, Gerber, H.-D, Garcia, G.A, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2002-10-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Virtual screening for submicromolar leads of tRNA-guanine transglycosylase based on a new unexpected binding mode detected by crystal structure analysis
J.Med.Chem., 46, 2003
|
|
6AZQ
 
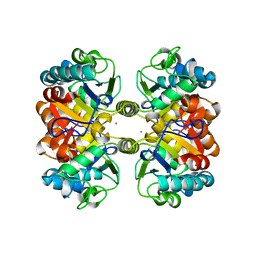 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CALCIUM ION, Putative amidase, dicarbonimidic diamide | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
1MVJ
 
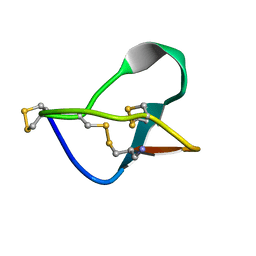 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1N41
 
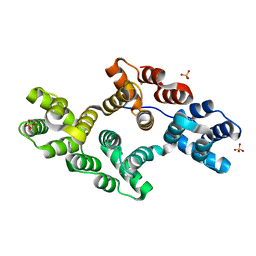 | | Crystal Structure of Annexin V K27E Mutant | | Descriptor: | CALCIUM ION, SULFATE ION, annexin V | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial basic cluster in annexin V couples phospholipid
binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
6AXA
 
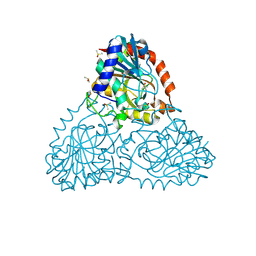 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with (3S)-5-fluoro-3-hydroxy-1,3-dihydroindol-2-one | | Descriptor: | (3S)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-fluoro-3-hydroxy-1,3-dihydroindol-2-one
To Be Published
|
|
4Z0W
 
 | | Peptaibol gichigamin isolated from Tolypocladium sup_5 | | Descriptor: | PEPTAIBOL GICHIGAMIN | | Authors: | Du, L, Risinger, A.L, Mitchell, C.A, Stamps, B.W, Pan, N, King, J.B, Motley, J.L, Thomas, L.M, Yang, Z, Stevenson, B.S, Mooberry, S.L, Cichewicz, R.H. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptaibol gichigamin isolated from Tolypocladium sup_5
TO BE PUBLISHED
|
|
1MUE
 
 | | Thrombin-Hirugen-L405,426 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDO-2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUOROPHENYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Nantermet, P.G, Selnick, H.G, Isaacs, R.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Stranieri, M.T, Cook, J.J, McMasters, D.R, Pellicore, J.M, Pal, S, Wallace, A.A, Clayton, F.C, Bohn, D, Welsh, D.C, Lynch, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacokinetic optimization of 3-amino-6-chloropyrazinone acetamide thrombin inhibitors. Implementation of P3 pyridine N-oxides to deliver an orally bioavailable series containing P1 N-benzylamides.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6ZW1
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | Descriptor: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | Authors: | Barinka, C, Motlova, L, Ustinova, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
1MVF
 
 | | MazE addiction antidote | | Descriptor: | PemI-like protein 1, immunoglobulin heavy chain variable region | | Authors: | Loris, R, Marianovsky, I, Lah, J, Laeremans, T, Engelberg-Kulka, H, Glaser, G, Muyldermans, S, Wyns, L. | | Deposit date: | 2002-09-25 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the intrinsically flexible addiction antidote MazE.
J.Biol.Chem., 278, 2003
|
|
1CJA
 
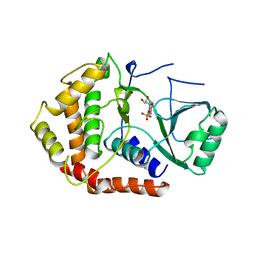 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
6B56
 
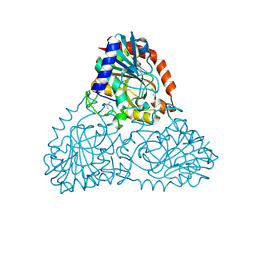 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-butylpyridine-2-carboxylic acid | | Descriptor: | 5-butylpyridine-2-carboxylic acid, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-28 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-butylpyridine-2-carboxylic acid
To Be Published
|
|
1MUO
 
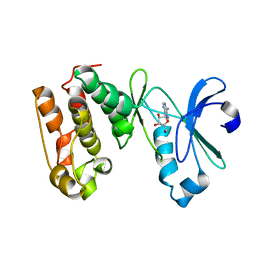 | | CRYSTAL STRUCTURE OF AURORA-2, AN ONCOGENIC SERINE-THREONINE KINASE | | Descriptor: | ADENOSINE, Aurora-related kinase 1 | | Authors: | Cheetham, G.M.T, Knegtel, R.M.A, Coll, J.T, Renwick, S.B, Swenson, L, Weber, P, Lippke, J.A, Austen, D.A. | | Deposit date: | 2002-09-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Aurora-2, an Oncogenic Serine/Threonine Kinase
J.Biol.Chem., 277, 2002
|
|
6B2L
 
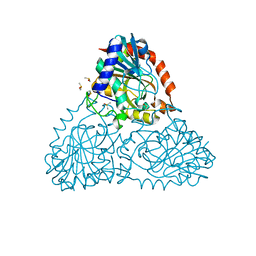 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with piperidin-2-Imine | | Descriptor: | DIMETHYL SULFOXIDE, PIPERIDIN-2-IMINE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with piperidin-2-Imine
To Be Published
|
|
1N44
 
 | | Crystal Structure of Annexin V R23E Mutant | | Descriptor: | Annexin V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interfacial basic cluster in anexin V couples phospholipid binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
4ZDS
 
 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
1N3H
 
 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | Descriptor: | SHC Transforming protein | | Authors: | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | Deposit date: | 2002-10-28 | | Release date: | 2003-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
7AAR
 
 | | sugar/H+ symporter STP10 in inward open conformation | | Descriptor: | CHLORIDE ION, Octyl Glucose Neopentyl Glycol, Sugar transport protein 10, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
