1W3M
 
 | | Crystal structure of tsushimycin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Delta-3isotetradecenoic acid, ... | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-07-16 | | Release date: | 2005-07-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the lipopeptide antibiotic tsushimycin.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
1W2U
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED THIO CELLOTETRAOSE | | Descriptor: | ENDOGLUCANASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1SDJ
 
 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1W6I
 
 | | plasmepsin II-pepstatin A complex | | Descriptor: | PEPSTATIN, PLASMEPSIN 2 PRECURSOR | | Authors: | Lindberg, J, Johansson, P.-O, Rosenquist, A, Kvarnstroem, I, Vrang, L, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-18 | | Release date: | 2006-07-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Study of a Novel Inhibitor with Bulky P1 Side Chain in Complex with Plasmepsin II -Implications for Drug Design
To be Published
|
|
1W18
 
 | | Crystal Structure of levansucrase from Gluconacetobacter diazotrophicus | | Descriptor: | LEVANSUCRASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Pons, T, Tarbouriech, N, Taylor, E.J, Hernandez, L, Davies, G.J. | | Deposit date: | 2004-06-16 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram- Negative Bacterium Gluconacetobacter Diazotrophicus.
Biochem.J., 390, 2005
|
|
5E9C
 
 | | Crystal structure of human heparanase in complex with heparin tetrasaccharide dp4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E4O
 
 | | Human transthyretin (TTR) complexed with (Z)-((3,4-Dichloro-phenyl)-methyleneaminooxy)-acetic acid | | Descriptor: | ({(Z)-[(3,4-dichlorophenyl)(phenyl)methylidene]amino}oxy)acetic acid, Transthyretin | | Authors: | Ciccone, L, Savko, M, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
8PKU
 
 | | Kelch domain of KEAP1 in complex with ortho-dimethylbenzene linked cyclic peptide 3 (ortho-WRCDEETGEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PKW
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 5 (ortho-WRCDPETaEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
5E70
 
 | | Crystal structure of Ecoli Branching Enzyme with gamma cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1S9F
 
 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
5EB0
 
 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5E86
 
 | | isolated SBD of BiP with loop34 modification | | Descriptor: | 78 kDa glucose-regulated protein | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
8PKX
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 11 (ortho-WRCNPETaEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PHE
 
 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
1YC0
 
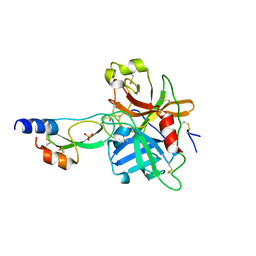 | | short form HGFA with first Kunitz domain from HAI-1 | | Descriptor: | Hepatocyte growth factor activator, Kunitz-type protease inhibitor 1, PHOSPHATE ION | | Authors: | Shia, S, Stamos, J, Kirchhofer, D, Fan, B, Wu, J, Corpuz, R.T, Santell, L, Lazarus, R.A, Eigenbrot, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B.
J.Mol.Biol., 346, 2005
|
|
1S6U
 
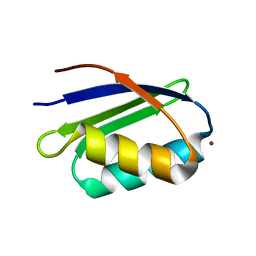 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1YKQ
 
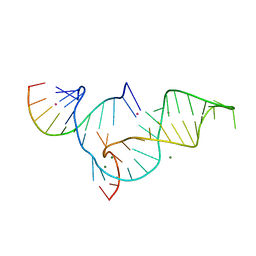 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7JHQ
 
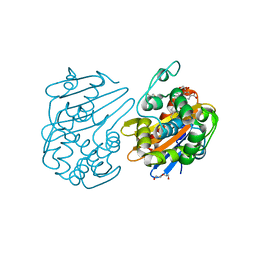 | | OXA-48 bound by Compound 2.3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-21 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
1YD2
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y19F bound to the catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
