1KE5
 
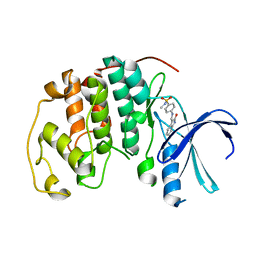 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1LJZ
 
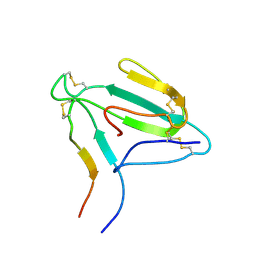 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1L4W
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Rodriguez, E, Eisenstein, M, Anglister, J. | | Deposit date: | 2002-03-06 | | Release date: | 2002-07-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1KE8
 
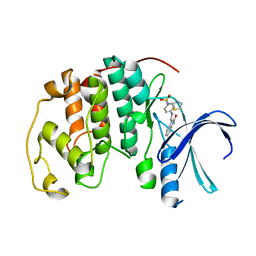 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE | | Descriptor: | 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE6
 
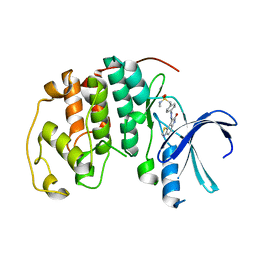 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1VT7
 
 | |
3UTZ
 
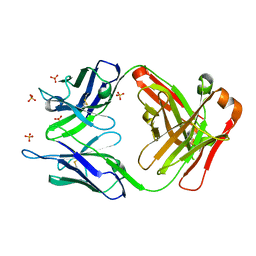 | | Endogenous-like inhibitory antibodies targeting activated metalloproteinase motifs show therapeutic potential | | Descriptor: | Metalloproteinase, heavy chain, light chain, ... | | Authors: | Sela-Passwell, N, Kikkeri, R, Dym, O, Rozenberg, H, Margalit, R, Arad-Yellin, R, Eisenstein, M, Brenner, O, Shoham, T, Danon, T, Shanzer, A, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-11-27 | | Release date: | 2011-12-14 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential.
NAT.MED. (N.Y.), 18, 2012
|
|
2F11
 
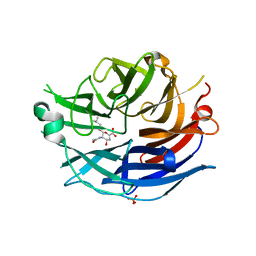 | | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor | | Descriptor: | 2-methylpropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor
To be Published
|
|
2F13
 
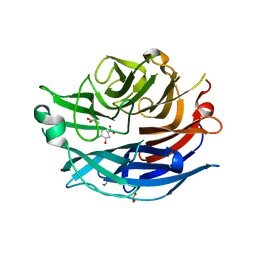 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor | | Descriptor: | (2R)-2,3-dihydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor
To be Published
|
|
2F12
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor | | Descriptor: | 3-hydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor
To be Published
|
|
4GB1
 
 | | Synthesis and Evaluation of Novel 3-C-alkylated-Neu5Ac2en Derivatives as Probes of Influenza Virus Sialidase 150-loop flexibility | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-phenylprop-2-en-1-yl]-D-glycero-L-altro-non-2-enonic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Rudrawar, S, Rameix-Welti, M.-A, Maggioni, A, Dyason, J.C, Rose, F.J, van der Werf, S, Thomson, R.J, Naffakh, N, Russell, R.J.M, von Itzstein, M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility.
Org.Biomol.Chem., 10, 2012
|
|
4FZG
 
 | | 20S yeast proteasome in complex with glidobactin | | Descriptor: | Glidobactin, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FZC
 
 | | 20S yeast proteasome in complex with cepafungin I | | Descriptor: | Cepafungin I, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LTC
 
 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
6QKX
 
 | |
6QKR
 
 | |
6QKG
 
 | | 2-Naphthoyl-CoA Reductase(NCR) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kayastha, K, Ermler, U. | | Deposit date: | 2019-01-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low potential enzymatic hydride transfer via highly cooperative and inversely functionalized flavin cofactors.
Nat Commun, 10, 2019
|
|
4IXD
 
 | | X-ray structure of lfa-1 i-domain in complex with ibe-667 at 1.8a resolution | | Descriptor: | 4-(3-{4-[(3-aminopropyl)carbamoyl]phenyl}-1H-indazol-1-yl)-N-methylbenzamide, Integrin alpha-L, MAGNESIUM ION | | Authors: | Kallen, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and x-ray structure based investigation of an ICAM-1 binding enhancing small molecule activator of LFA-1
To be Published, 2013
|
|
2P4Y
 
 | | Crystal structure of human PPAR-gamma-ligand binding domain complexed with an indole-based modulator | | Descriptor: | (2R)-2-(4-CHLORO-3-{[3-(6-METHOXY-1,2-BENZISOXAZOL-3-YL)-2-METHYL-6-(TRIFLUOROMETHOXY)-1H-INDOL-1-YL]METHYL}PHENOXY)PROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | McKeever, B.M. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The differential interactions of peroxisome proliferator-activated receptor gamma ligands with Tyr473 is a physical basis for their unique biological activities.
Mol.Pharmacol., 73, 2008
|
|
8D3A
 
 | | Crystal Structure of DH475 Fab in complex with Man9 | | Descriptor: | DH475 Fab heavy chain, DH475 Fab light chain, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Finkelstein, M, Fera, D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of two cooperating antibodies unveils immune pressure imposed on HIV Env to elicit a V3-glycan supersite broadly neutralizing antibody lineage.
Front Immunol, 13, 2022
|
|
1EP8
 
 | | CRYSTAL STRUCTURE OF A MUTATED THIOREDOXIN, D30A, FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
1FCT
 
 | |
4R02
 
 | | yCP in complex with BSc4999 (alpha-Keto Phenylamide) | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(2S,3S)-1-[(2,4-dimethylphenyl)amino]-2-hydroxy-5-methyl-1-oxohexan-3-yl}-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Voss, C, Scholz, C, Knorr, S, Beck, P, Stein, M, Zall, A, Kuckelkorn, U, Kloetzel, P.-M, Groll, M, Hamacher, K, Schmidt, B. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | alpha-Keto Phenylamides as P1'-Extended Proteasome Inhibitors.
Chemmedchem, 9, 2014
|
|
1DBY
 
 | | NMR STRUCTURES OF CHLOROPLAST THIOREDOXIN M CH2 FROM THE GREEN ALGA CHLAMYDOMONAS REINHARDTII | | Descriptor: | CHLOROPLAST THIOREDOXIN M CH2 | | Authors: | Lancelin, J.-M, Guilhaudis, L, Krimm, I, Blackledge, M.J, Marion, D. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thioredoxin m from the green alga Chlamydomonas reinhardtii.
Proteins, 41, 2000
|
|
4HNP
 
 | | Crystal structure of yeast 20S proteasome in complex with vinylketone carmaphycin analogue VNK1 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylhept-1-en-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-20 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
