5HHD
 
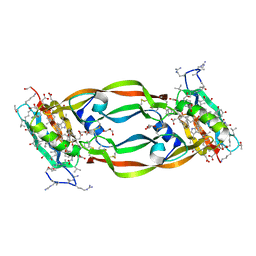 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5HHC
 
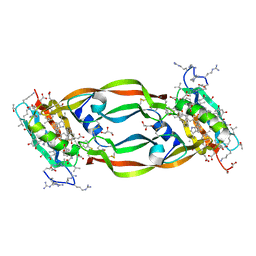 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
4GLN
 
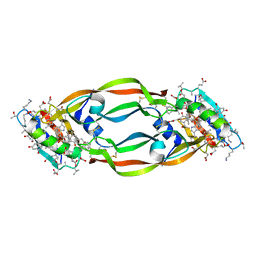 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D-RFX001, Vascular endothelial growth factor A | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLS
 
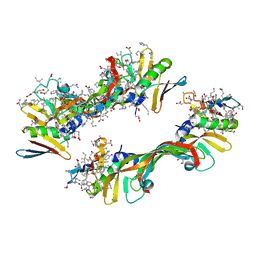 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6EK2
 
 | |
3N85
 
 | |
6IKM
 
 | | Crystal structure of SpuE-Spermidine in complex with ScFv5 | | Descriptor: | Polyamine transport protein, SPERMIDINE, SULFATE ION, ... | | Authors: | Wu, D, Sun, X. | | Deposit date: | 2018-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A Potent Anti-SpuE Antibody Allosterically Inhibits Type III Secretion System and Attenuates Virulence of Pseudomonas Aeruginosa.
J.Mol.Biol., 431, 2019
|
|
2P3W
 
 | |
7M6T
 
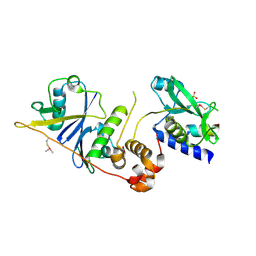 | | Crystal structure of SOCS2/ElonginB/ElonginC bound to a non-canonical peptide that enhances phospho-peptide binding | | Descriptor: | Elongin-B, Elongin-C, Non-canonical peptide F3, ... | | Authors: | Kershaw, N.J, Li, K, Linossi, E.M, Nicholson, S.E. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.
Nat Commun, 12, 2021
|
|
4Q2P
 
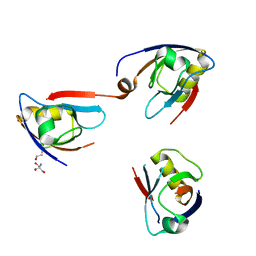 | | NHERF3 PDZ2 in Complex with a Phage-Derived Peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2014-04-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural portrait of the PDZ domain family.
J.Mol.Biol., 426, 2014
|
|
4Q2N
 
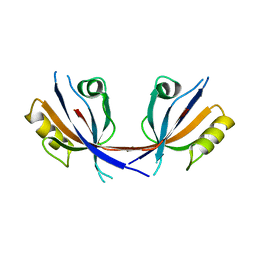 | |
6D4P
 
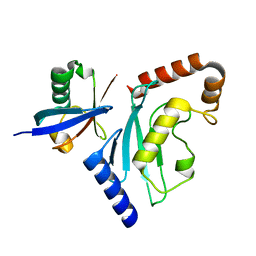 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D68
 
 | | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1, Ubv.G1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D6I
 
 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
5C7J
 
 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
3P9W
 
 | |
8CAF
 
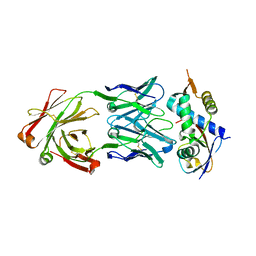 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6B57
 
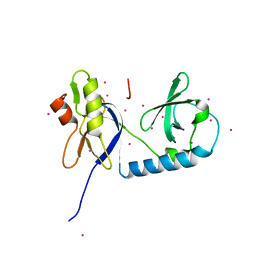 | | tudor in complex with ligand | | Descriptor: | Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6OB1
 
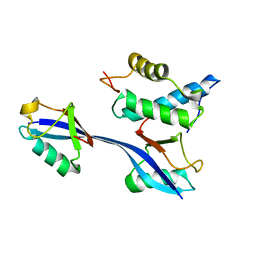 | | Structure of WHB in complex with Ubiquitin Variant | | Descriptor: | Anaphase-promoting complex subunit 2, Ubiquitin | | Authors: | Edmond, R.W, Grace, C.R. | | Deposit date: | 2019-03-19 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QK9
 
 | |
7Q51
 
 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
 | | human Gid4 bound to a Phe/N-peptide | | Descriptor: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | Authors: | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
