6D60
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVS
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.318 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D61
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6MSM
 
 | | Phosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the ATP-bound, phosphorylated human CFTR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1XAX
 
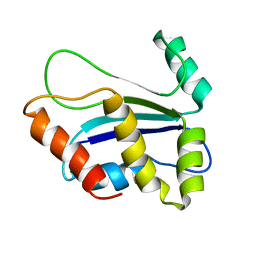 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
4NPI
 
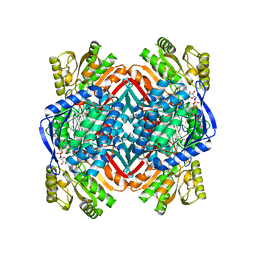 | | 1.94 Angstroms X-ray crystal structure of NAD- and intermediate- bound alpha-aminomuconate-epsilon-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4OFC
 
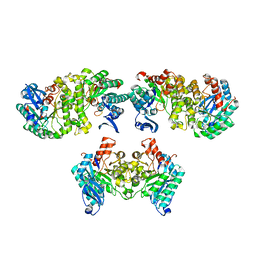 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
4OE2
 
 | | 2.00 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Esaki, S, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4X80
 
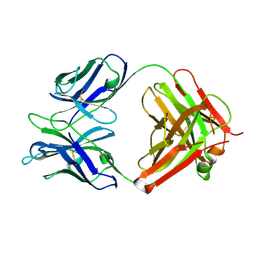 | | Crystal Structure of murine 7B4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | IgG1 7B4 FAB Heavy chain, IgG1 7B4 FAB Light Chain | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
4X8J
 
 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
3OKX
 
 | | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris | | Descriptor: | S-ADENOSYLMETHIONINE, YaeB-like protein RPA0152 | | Authors: | Chang, C, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris
To be Published
|
|
3PF6
 
 | | The structure of uncharacterized protein PP-LUZ7_gp033 from Pseudomonas phage LUZ7. | | Descriptor: | CHLORIDE ION, hypothetical protein PP-LUZ7_gp033 | | Authors: | Cuff, M.E, Evdokimova, E, Liu, F, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of uncharacterized protein PP-LUZ7_gp033 from Pseudomonas phage LUZ7.
TO BE PUBLISHED
|
|
2AOF
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | Descriptor: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2ELA
 
 | | Crystal Structure of the PTB domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2ELB
 
 | | Crystal Structure of the BAR-PH domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2AOE
 
 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | Descriptor: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
8JRE
 
 | | Cryo-EM structure of a designed AAV8-based vector | | Descriptor: | Capsid protein | | Authors: | Ke, X, Luo, S, Zheng, Q, Jiang, H, Liu, F, Sun, X. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | An adeno-associated virus variant enabling efficient ocular-directed gene delivery across species.
Nat Commun, 15, 2024
|
|
2AOJ
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P6-PR | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOC
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOD
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
5UDY
 
 | | Human alkaline sphingomyelinase (alk-SMase, ENPP7, NPP7) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Liu, F, Illes, K, Nagar, B. | | Deposit date: | 2016-12-28 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition.
J. Biol. Chem., 292, 2017
|
|
