4QUV
 
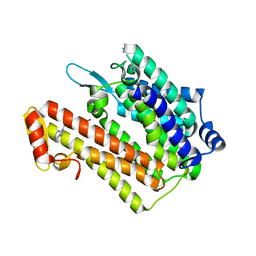 | | Structure of an integral membrane delta(14)-sterol reductase | | Descriptor: | Delta(14)-sterol reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, X, Blobel, G. | | Deposit date: | 2014-07-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum.
Nature, 517, 2015
|
|
4QRD
 
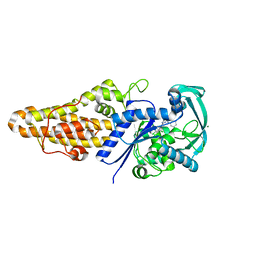 | | Structure of Methionyl-tRNA Synthetase in complex with N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Descriptor: | MAGNESIUM ION, Methionyl-tRNA synthetase, N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Authors: | Li, X, Hilgers, M.T, Stidham, M, Brown-Driver, V, Shaw, K.J, Finn, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and SAR of a Novel Series of Pyrimidine Antibacterials Targeting Methionyl-tRNA Synthetase
to be published
|
|
5U74
 
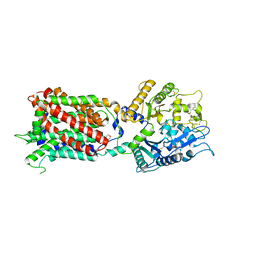 | | Structure of human Niemann-Pick C1 protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VB3
 
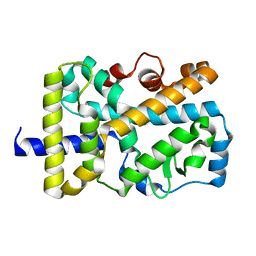 | | X-ray structure of nuclear receptor ROR-gammat Ligand Binding Domain + SRC2 peptide | | Descriptor: | Nuclear receptor ROR-gamma, SRC2 chimera, SODIUM ION | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5V5L
 
 | |
5VB6
 
 | |
5VB5
 
 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an inverse agonist and SRC2 peptide | | Descriptor: | N-[(2R)-3-(4-{[3-(4-chlorophenyl)propanoyl]amino}phenyl)-1-(4-methylpiperidin-1-yl)-1-oxopropan-2-yl]-4-methylpentanamide, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5VQK
 
 | |
5VQL
 
 | |
5WNE
 
 | |
5WNG
 
 | |
5WNH
 
 | |
5WNF
 
 | | X-RAY CO-STRUCTURE OF RHO-ASSOCIATED PROTEIN KINASE (ROCK1) WITH A HIGHLY SELECTIVE INHIBITOR | | Descriptor: | 1-(4-amino-1,2,5-oxadiazol-3-yl)-5-methyl-N-({3-[(5-methyl-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl)carbamoyl]phenyl}methyl)-1H-1,2,3-triazole-4-carboxamide, GLYCEROL, Rho-associated protein kinase 1 | | Authors: | Li, X. | | Deposit date: | 2017-07-31 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel mechanism of Rho kinase selectivity: beyond the ATP pocket
To Be Published
|
|
4JPB
 
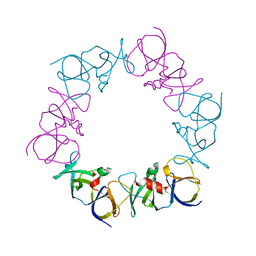 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
4KMN
 
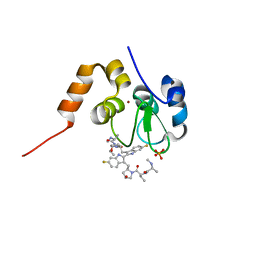 | | Structure of cIAP1-BIR3 and inhibitor | | Descriptor: | (2S)-N-{(2R)-1-[(2R,4S)-2-{[6,6'-difluoro-3'-({(2R,4S)-4-hydroxy-1-[(2S)-2-{[(2S)-2-(methylamino)propanoyl]amino}butanoyl]pyrrolidin-2-yl}methyl)-1H,1'H-2,2'-biindol-3-yl]methyl}-4-hydroxypyrrolidin-1-yl]-1-oxobutan-2-yl}-2-(methylamino)propanamide, Baculoviral IAP repeat-containing protein 2, PHOSPHATE ION, ... | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of cIAP1-BIR3 and inhibitor
To be Published
|
|
6A5M
 
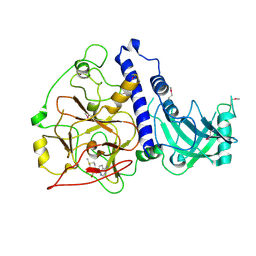 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5V5M
 
 | |
5VB7
 
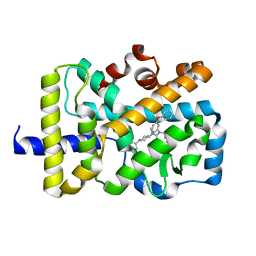 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an agonist and SRC2 peptide | | Descriptor: | N-methyl-N'-(3-methylbut-2-en-1-yl)-N'-(3-phenoxyphenyl)-N-[trans-4-(pyridin-4-yl)cyclohexyl]urea, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
6A5N
 
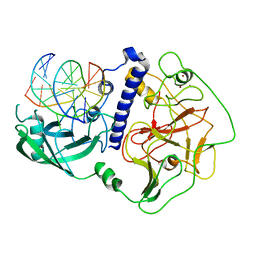 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
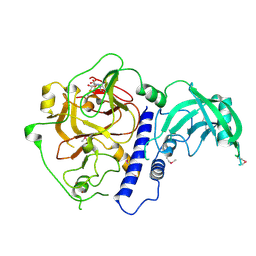 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4KMP
 
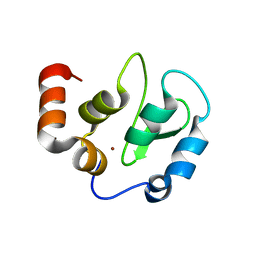 | | Structure of XIAP-BIR3 and inhibitor | | Descriptor: | (2S,2'S)-N,N'-[(6,6'-difluoro-1H,1'H-2,2'-biindole-3,3'-diyl)bis{methanediyl[(2R,4S)-4-hydroxypyrrolidine-2,1-diyl][(2S)-1-oxobutane-1,2-diyl]}]bis[2-(methylamino)propanamide], E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of XIAP-BIR3 and inhibitor
To be Published
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2B
 
 | | Cryo-EM structure of the human 39S mitoribosome with Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2A
 
 | | Cryo-EM structure of the human 55S mitoribosome with Tigecycline | | Descriptor: | 12S rRNA, 16S rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XWD
 
 | | Croy-EM structure of alpha synuclein fibril with EGCG | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Alpha-synuclein | | Authors: | Li, X, Liu, C. | | Deposit date: | 2024-01-16 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single-Molecule Insight Into alpha-Synuclein Fibril Structure and Mechanics Modulated by Chemical Compounds.
Adv Sci, 12, 2025
|
|
