6JMB
 
 | |
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
8IYS
 
 | | TUG891-bound FFAR4 in complex with Gq | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
6L8L
 
 | | C-Src in complex with ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Characterization of ibrutinib as a non-covalent inhibitor of SRC-family kinases.
Bioorg.Med.Chem.Lett., 34, 2020
|
|
8H5T
 
 | |
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
8H4I
 
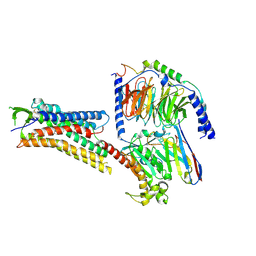 | | DHA-bound FFAR4 in complex with Gs | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4L
 
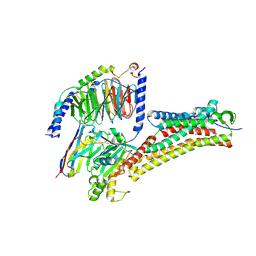 | | DHA-bound FFAR4 in complex with Gq | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4K
 
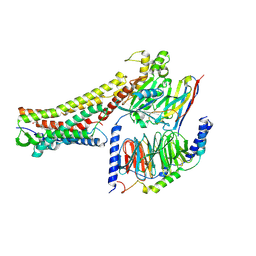 | | GW9508-bound FFAR4 in complex with Gq | | Descriptor: | 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
7XXJ
 
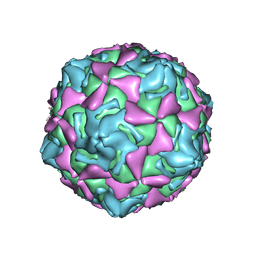 | | Echo 18 incubated with FcRn at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXA
 
 | | Complex of Echo 18 and FcRn at pH7.4 | | Descriptor: | IgG receptor FcRn large subunit p51, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
