7ZAY
 
 | | Human heparan sulfate polymerase complex EXT1-EXT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-1, Exostosin-2, ... | | Authors: | Leisico, F, Omeiri, J, Hons, M, Schoehn, G, Lortat-Jacob, H, Wild, R. | | Deposit date: | 2022-03-23 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human heparan sulfate polymerase complex EXT1-EXT2.
Nat Commun, 13, 2022
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | Descriptor: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7N03
 
 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
6O9C
 
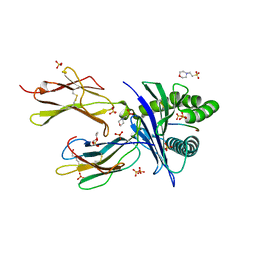 | |
4TRO
 
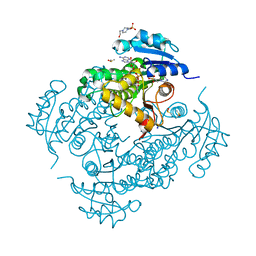 | | Structure of the enoyl-ACP reductase of Mycobacterium tuberculosis InhA, inhibited with the active metabolite of isoniazid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
4TRM
 
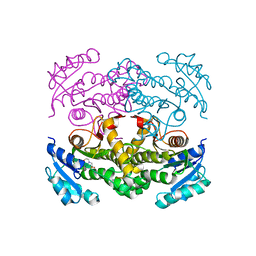 | | Structure of the apo form of InhA from Mycobacterium tuberculosis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
6O9B
 
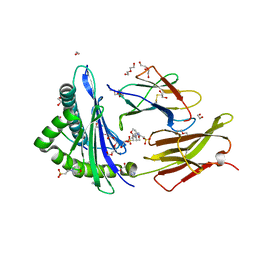 | |
4TRN
 
 | | STRUCTURE OF INHA FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED TO NADH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, INHA, ... | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
7STF
 
 | | Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Wright, K.M, Gabelli, S.B, Miller, M. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
8FMX
 
 | | Langya virus F glycoprotein ectodomain in prefusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Low, Y.S, Isaacs, A, Modhiran, N, Watterson, D. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure and antigenicity of divergent Henipavirus fusion glycoproteins.
Nat Commun, 14, 2023
|
|
8FMY
 
 | |
6JD7
 
 | | Crystal structure of anti-CRISPR protein AcrIIC2 dimer | | Descriptor: | ACETATE ION, AcrIIC2, MAGNESIUM ION | | Authors: | Cheng, Z, Huang, X, Sun, W, Wang, Y.L. | | Deposit date: | 2019-01-31 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6JDX
 
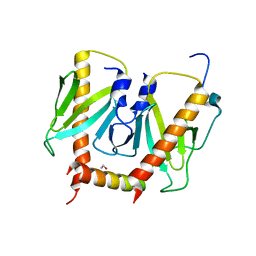 | | Crystal structure of AcrIIC2 dimer in complex with partial Nme1Cas9 preprocessed with protease alpha-Chymotrypsin | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Cheng, Z, Huang, X, Sun, W, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6JDJ
 
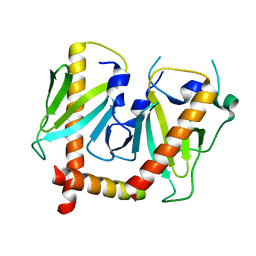 | | Crystal structure of AcrIIC2 dimer in complex with partial Nme1Cas9 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Cheng, Z, Huang, X, Sun, W, Wang, Y. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6H7X
 
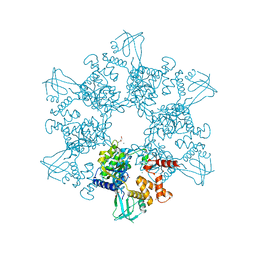 | | First X-ray structure of full-length human RuvB-Like 2. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Silva, S, Brito, J.A, Matias, P, Bandeiras, T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | X-ray structure of full-length human RuvB-Like 2 - mechanistic insights into coupling between ATP binding and mechanical action.
Sci Rep, 8, 2018
|
|
1S17
 
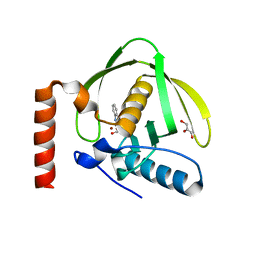 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6N05
 
 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
8G0G
 
 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
8G0F
 
 | |
5OL0
 
 | | Structure of Leishmania infantum Silent Information Regulator 2 related protein 1 (LiSIR2rp1) in complex with acetylated p53 peptide | | Descriptor: | Cellular tumor antigen p53, Putative silent information regulator 2,Putative silent information regulator 2, ZINC ION | | Authors: | Ronin, C, Ciesielski, F, Ciapetti, P. | | Deposit date: | 2017-07-26 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the Leishmania infantum Silent Information Regulator 2 related protein 1: Implications to protein function and drug design.
PLoS ONE, 13, 2018
|
|
6GP0
 
 | | Structure of mEos4b in the red fluorescent state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
6GP1
 
 | | Structure of mEos4b in the red long-lived dark state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
6GOY
 
 | | Structure of mEos4b in the green fluorescent state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
