7JQN
 
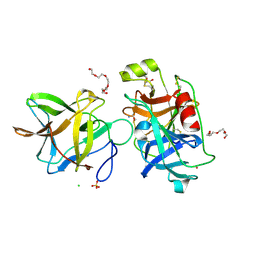 | | Crystal structure of the R64M mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JR2
 
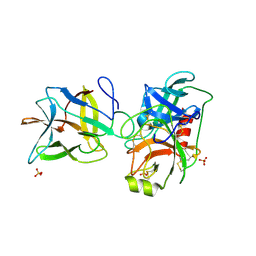 | |
7JR1
 
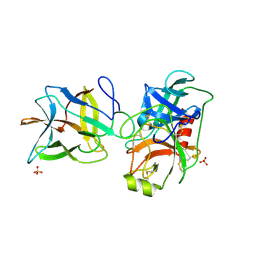 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Bovine Trypsin | | Descriptor: | Cationic trypsin, Kunitz-type inihibitor, SODIUM ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JQK
 
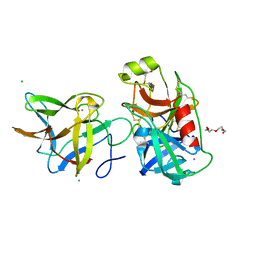 | | Crystal structure of the R64A mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JQV
 
 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CHLORIDE ION, Kallikrein 4 (Prostase, enamel matrix, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3ZMZ
 
 | | LSD1-CoREST in complex with PRSFAV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
5H2I
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 110 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
1SHR
 
 | | Crystal structure of ferrocyanide bound human hemoglobin A2 at 1.88A resolution | | Descriptor: | CYANIDE ION, FE (III) ION, Hemoglobin alpha chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb
Biochemistry, 43, 2004
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7BD7
 
 | |
7BRD
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Mundra, S, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural and functional characterization of peptidyl-tRNA hydrolase from Klebsiella pneumoniae.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
2PD1
 
 | | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea | | Descriptor: | ACETATE ION, Hypothetical protein | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea.
To be Published
|
|
1KWD
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CONSERVED REGION OF HUMAN RESPIRATORY SYNCYTIAL VIRUS ATTACHMENT GLYCOPROTEIN G 187 | | Descriptor: | MAJOR SURFACE GLYCOPROTEIN G | | Authors: | Sugawara, M, Czaplicki, J, Ferrage, J, Haeuw, J.F, Power, U.F, Corvaia, N, Nguyen, T, Beck, A, Milon, A. | | Deposit date: | 2002-01-29 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G.
J.Pept.Res., 60, 2002
|
|
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
5B6Y
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 36.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-15 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
2OZV
 
 | | Crystal structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens. | | Descriptor: | Hypothetical protein Atu0636 | | Authors: | Cuff, M.E, Xu, X, Zheng, X, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
6R6F
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
1KWE
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CONSERVED REGION OF HUMAN RESPIRATORY SYNCYTIAL VIRUS ATTACHMENT GLYCOPROTEIN G | | Descriptor: | MAJOR SURFACE GLYCOPROTEIN G | | Authors: | Sugawara, M, Czaplicki, J, Ferrage, J, Haeuw, J.F, Power, U.F, Corvaia, N, Nguyen, T, Beck, A, Milon, A. | | Deposit date: | 2002-01-29 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G.
J.Pept.Res., 60, 2002
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5AX2
 
 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
3I4C
 
 | | Crystal structure of Sulfolobus Solfataricus ADH(SsADH) double mutant (W95L,N249Y) | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Pennacchio, A, Zagari, A, Rossi, M, Raia, C.A. | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase
Extremophiles, 13, 2009
|
|
1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
5EGR
 
 | | tRNA guanine transglycosylase (TGT) in complex with an Immucillin derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-7-[(2~{S},3~{R},5~{S})-5-(hydroxymethyl)-3-oxidanyl-pyrrolidin-2-yl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of an Immucillin Derivative as a Class of tRNA-Guanine Transglycosylase Inhibitors: Exploration of a Transition State Analogous Binding Mode
To be Published
|
|
2GRG
 
 | | Solution NMR Structure of Protein YNR034W-A from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YT727; Ontario Center for Structural Proteomics Target yst6499. | | Descriptor: | hypothetical protein; Ynr034w-ap | | Authors: | Wu, B, Yee, A, Ramelot, T, Lemak, A, Semesi, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein yst6499 from Saccharomyces cerevisiae/ Northeast Structural Genomics Consortium Target YT727/ Ontario Center for Structural Proteomics Target yst6499
To be Published
|
|
