1UJP
 
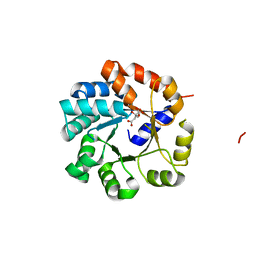 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, Tryptophan synthase alpha chain | | Authors: | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-08 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
8T9F
 
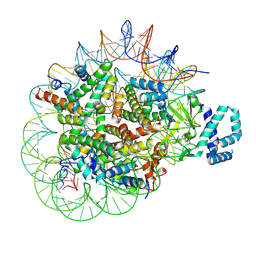 | |
8SXG
 
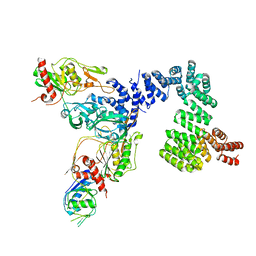 | |
8SXE
 
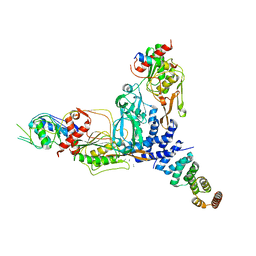 | |
8SXH
 
 | |
8SXF
 
 | |
5EUV
 
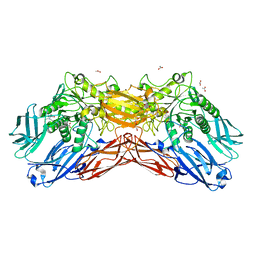 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1QO2
 
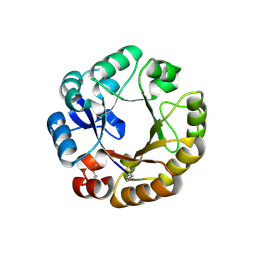 | | Crystal structure of N-((5'-phosphoribosyl)-formimino)-5-aminoimidazol-4-carboxamid ribonucleotid isomerase (EC 3.1.3.15, HisA) | | Descriptor: | | | Authors: | Wilmanns, M, Lang, D, Thoma, R, Sterner, R. | | Deposit date: | 1999-11-01 | | Release date: | 2000-07-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Evidence for Evolution of the Beta/Alpha-Barrel Scaffold by Repeated Gene Duplication and Fusion
Science, 289, 2000
|
|
8DNN
 
 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
8THU
 
 | |
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
6LR6
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
6LPF
 
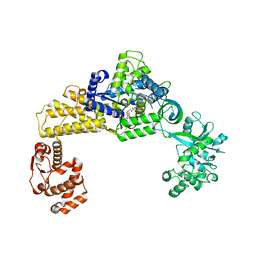 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
5NGE
 
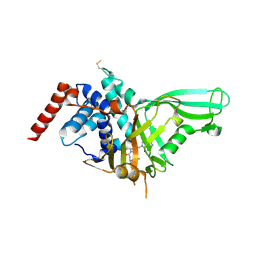 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
7BPC
 
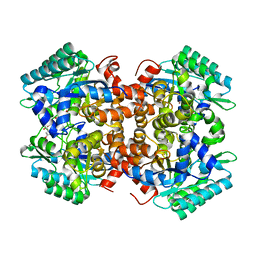 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7E1L
 
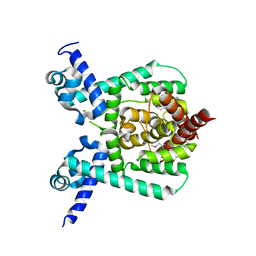 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
5H6X
 
 | | The crystal structure of RpoS fragment including a partial region 1.2 and region 2 from intracellular pathogen Legionella pneumophila | | Descriptor: | GLYCEROL, RNA polymerase sigma factor RpoS, SODIUM ION | | Authors: | Zhang, N, Chen, X, Gong, X, Ge, H. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of RpoS Fragment including a Partial Region 1.2 and Region 2 from the Intracellular Pathogen Legionella pneumophila
Crystals, 8, 2018
|
|
