3GWB
 
 | | Crystal structure of peptidase M16 inactive domain from Pseudomonas fluorescens. Northeast Structural Genomics target PlR293L | | Descriptor: | Peptidase M16 inactive domain family protein | | Authors: | Seetharaman, J, Chen, Y, Fang, F, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptidase M16 inactive domain from Pseudomonas fluorescens. Northeast Structural Genomics target PlR293L
To be Published
|
|
3EVI
 
 | | Crystal structure of the thioredoxin-fold domain of human phosducin-like protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosducin-like protein 2, SODIUM ION | | Authors: | Lou, X, Bao, R, Zhou, C, Chen, Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the thioredoxin-fold domain of human phosducin-like protein 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3ESH
 
 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | Descriptor: | Protein similar to metal-dependent hydrolase | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
3GQQ
 
 | | Crystal structure of the human retinal protein 4 (unc-119 homolog A). Northeast Structural Genomics Consortium target HR3066a | | Descriptor: | Protein unc-119 homolog A, UNKNOWN LIGAND | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Crystal structure of the human retinal protein 4 (unc-119 homolog A).
To be Published
|
|
8W9Y
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5XP5
 
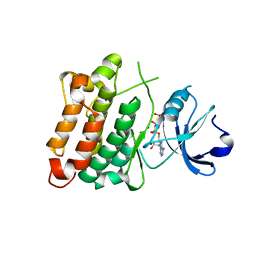 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
2LVN
 
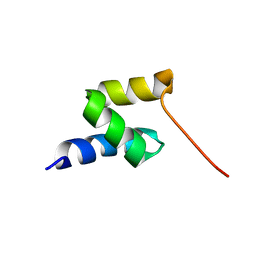 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVO
 
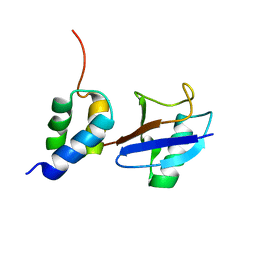 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVQ
 
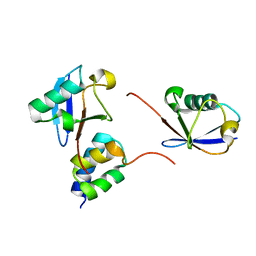 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
6B0V
 
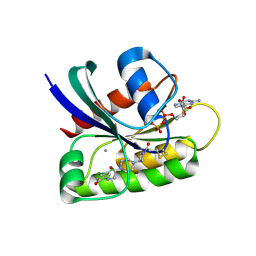 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2O3I
 
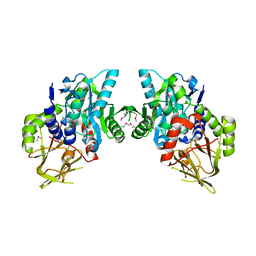 | | X-ray Crystal Structure of Protein CV_3147 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR68. | | Descriptor: | Hypothetical protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Cunningham, K, Ma, L.C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the hypothetical protein Q7NTB2_CHRVO from Chromobacterium violaceum
To be Published
|
|
2P0V
 
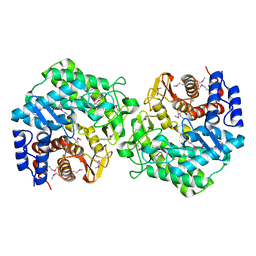 | | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 | | Descriptor: | Hypothetical protein BT3781 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-20 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
2OA5
 
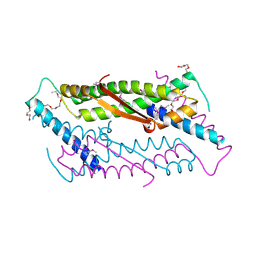 | | Crystal structure of ORF52 from Murid herpesvirus (MUHV-4) (Murine gammaherpesvirus 68) at 2.1 A resolution. Northeast Structural Genomics Consortium target MHR28B. | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68.
J.Biol.Chem., 282, 2007
|
|
5YRP
 
 | | Crystal structure of the EAL domain of Mycobacterium smegmatis DcpA | | Descriptor: | MAGNESIUM ION, Sensory box/response regulator | | Authors: | Chen, H.J, li, N, Luo, Y, Jiang, Y.L, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP inMycobacterium smegmatis.
Biochem. J., 475, 2018
|
|
2PX9
 
 | | The intrinsic affinity between E2 and the Cys domain of E1 in Ubiquitin-like modifications | | Descriptor: | SUMO-activating enzyme subunit 2, SUMO-conjugating enzyme UBC9 | | Authors: | Wang, J.H, Hu, W.D, Cai, S, Lee, B, Song, J, Chen, Y. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications.
Mol.Cell, 27, 2007
|
|
5XP7
 
 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
2PIG
 
 | | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution. Northeast Structural Genomics target SCR6 | | Descriptor: | Putative transferase, ZINC ION | | Authors: | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Ho, C.K, Janjua, H, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution.
To be Published
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2P0Y
 
 | | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR6 | | Descriptor: | Hypothetical protein lp_0780 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Chi, K.H, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum.
To be Published
|
|
2QYK
 
 | | Crystal structure of PDE4A10 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, Cyclic AMP-specific phosphodiesterase HSPDE4A10, MAGNESIUM ION, ... | | Authors: | Wang, H, Peng, M, Chen, Y, Geng, J, Robinson, H, Houslay, M. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
3LD7
 
 | | Crystal structure of the Lin0431 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Vorobiev, S, Chen, Y, Lee, D, Patel, D.J, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-12 | | Release date: | 2010-01-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Crystal structure of the Lin0431 protein from Listeria innocua
To be Published
|
|
3LD0
 
 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
