7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YRZ
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, Y.R, Zeng, P, Zhou, X.L, Lin, C, Zhang, J, Yin, X.S, Li, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
8SNY
 
 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8SNX
 
 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the leader promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*GP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
8TU6
 
 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSA
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J, Valverde, R. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS8
 
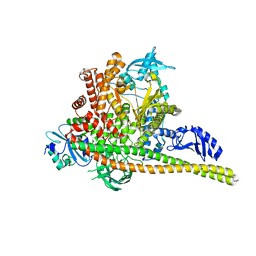 | | p85alpha/p110alpha heterodimer H1047R mutant | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSB
 
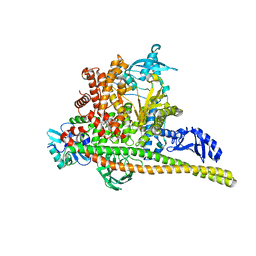 | | Human PI3K p85alpha/p110alpha bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSD
 
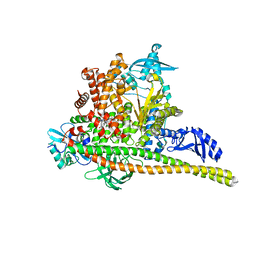 | | Human PI3K p85alpha/p110alpha bound to RLY-2608 | | Descriptor: | N-{(3R,6M)-3-(2-chloro-5-fluorophenyl)-6-[(4S)-5-cyano[1,2,4]triazolo[1,5-a]pyridin-6-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-3-fluoro-5-(trifluoromethyl)benzamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS7
 
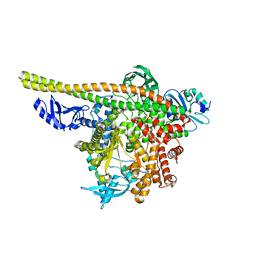 | | Human PI3K p85alpha/p110alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSC
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 3 | | Descriptor: | (1S)-7-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-1-(2-methylphenyl)-3-oxo-2,3-dihydro-1H-isoindole-5-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS9
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 1 | | Descriptor: | 5-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7WSF
 
 | |
