4NB6
 
 | | Crystal structure of the ligand binding domain of RORC with T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Hymowitz, S.G, Boenig-de Leon, G. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of substituted hexafluoroisopropanol-arylsulfonamides as modulators of RORc.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5GIM
 
 | | Crystal structure of thrombin-avathrin complex | | Descriptor: | C-terminal peptide from Putative uncharacterized protein avahiru, N-terminal peptide from Putative uncharacterized protein avahiru, Thrombin light chain, ... | | Authors: | Kini, R.M, Koh, C.Y, Iyer, J.K, Swaminathan, K. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Avathrin: a novel thrombin inhibitor derived from a multicopy precursor in the salivary glands of the ixodid tick, Amblyomma variegatum.
FASEB J., 31, 2017
|
|
8I54
 
 | | Lb2Cas12a RNA DNA complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3'), Lb2Cas12a, ... | | Authors: | Li, J, Sivaraman, J, Satoru, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
6INU
 
 | |
6INN
 
 | |
6INV
 
 | |
5UTY
 
 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
7SWD
 
 | | Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound | | Descriptor: | 1C11 scFv, 1C3 heavy chain, 1C3 light chain, ... | | Authors: | Milligan, J.C, Yu, X, Saphire, E.O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
6INO
 
 | |
8K8H
 
 | |
6IOE
 
 | |
9BLX
 
 | | Rhesus macaque ITS111.01 Fab in complex with SIV MPER peptide | | Descriptor: | Envelope glycoprotein gp160, GLYCEROL, ITS111.01 Heavy, ... | | Authors: | Gorman, J, Ahmadi, M, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Isolation and structure of broad SIV-neutralizing antibodies reveal a proximal helical MPER epitope recognized by a rhesus multi-donor class.
Cell Rep, 44, 2025
|
|
9BNS
 
 | |
9BP1
 
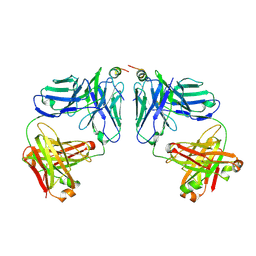 | | Rhesus macaque ITS110.01 Fab in complex with SIV Env MPER peptide | | Descriptor: | Envelope glycoprotein gp160, ITS110.01 Heavy Chain, ITS110.01 Light Chain | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-05-06 | | Release date: | 2025-01-15 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Isolation and structure of broad SIV-neutralizing antibodies reveal a proximal helical MPER epitope recognized by a rhesus multi-donor class.
Cell Rep, 44, 2025
|
|
5UTF
 
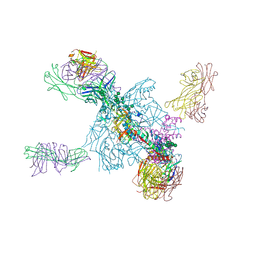 | | Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35022 Heavy chain, ... | | Authors: | Pancera, M, Chuang, G.-Y, Xu, K, Kwong, P.D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
7C00
 
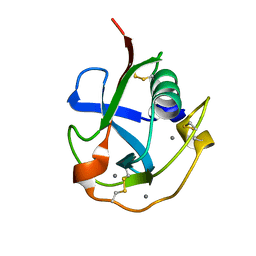 | | Crystal structure of the SRCR domain of human SCARA5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
6VZI
 
 | |
1DAN
 
 | |
3O7L
 
 | |
6W03
 
 | |
7BZZ
 
 | | Crystal structure of the SRCR domain of mouse SCARA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
3X11
 
 | | Crystal structure of HLA-B*57:01.I80N.L82R.R83G | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
3X13
 
 | | Crystal structure of HLA-B*0801.N80I | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
4M7R
 
 | |
5B39
 
 | | KIR3DL1*015 in complex with HLA-B*57:01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition
J.Exp.Med., 213, 2016
|
|
