3TFU
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, post-reaction complex with a 3,6-dihydropyrid-2-one heterocycle inhibitor | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DIMETHYL SULFOXIDE, [5-hydroxy-4-({[6-(3-hydroxypropyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Geders, T.W, Finzel, B.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism-based Inactivation by Aromatization of the Transaminase BioA Involved in Biotin Biosynthesis in Mycobaterium tuberculosis.
J.Am.Chem.Soc., 133, 2011
|
|
5I2E
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) with Bound Sulfamate Inhibitor 3a:3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine | | Descriptor: | 3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine, GLYCEROL, Histidine triad nucleotide-binding protein 1 | | Authors: | Strom, A.M, Finzel, B.C, Wagner, C.R. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
5IPC
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5I2F
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) with bound sulfamide inhibitor Bio-AMS | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
5IPE
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPD
 
 | |
5IPB
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
4CXR
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with 1-(1,3- benzothiazol-2-yl)methanamine | | Descriptor: | 1,2-ETHANEDIOL, 1-(1,3-benzothiazol-2-yl)methanamine, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Geders, T.W, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
4CXQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
4XTU
 
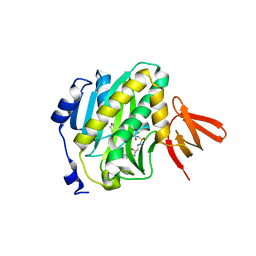 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
7SEO
 
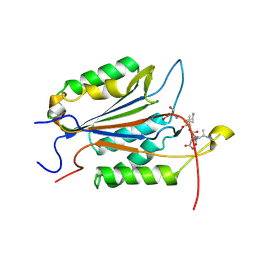 | | Crystal Structure of Caspase-3 with Peptide Inhibitor Ac-VDV(DAB)D-CHO | | Descriptor: | ACE-VAL-ASP-VAL-DAB-ASP, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fuller, J.L, Finzel, B.C. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
1RSM
 
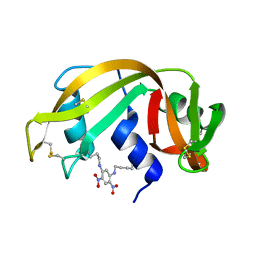 | | THE 2-ANGSTROMS RESOLUTION STRUCTURE OF A THERMOSTABLE RIBONUCLEASE A CHEMICALLY CROSS-LINKED BETWEEN LYSINE RESIDUES 7 AND 41 | | Descriptor: | DINITROPHENYLENE, RIBONUCLEASE A | | Authors: | Weber, P.C, Sheriff, S, Ohlendorf, D.H, Finzel, B.C, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2-A resolution structure of a thermostable ribonuclease A chemically cross-linked between lysine residues 7 and 41.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
7RNA
 
 | |
7RNG
 
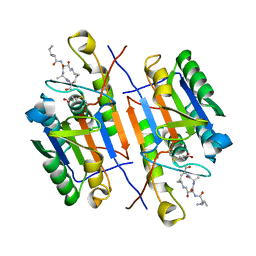 | |
5KGT
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead: 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone | | Descriptor: | 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Maize, K.M, Divakaran, A, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
5KGS
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead: 5-[4-(1,3-benzodioxol-5-ylcarbonyl)piperazin-1-yl]-2,3-dihydroinden-1-one | | Descriptor: | 5-[4-(1,3-benzodioxol-5-ylcarbonyl)piperazin-1-yl]-2,3-dihydroinden-1-one, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Maize, K.M, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
5KM4
 
 | |
5KLZ
 
 | |
5KM5
 
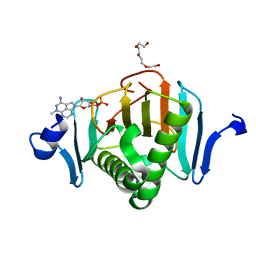 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) triciribine 5'-monoposphate catalytic product complex | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, CHLORIDE ION, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM9
 
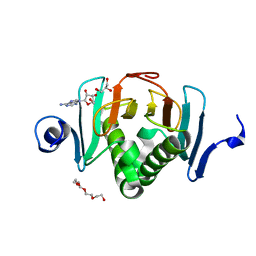 | |
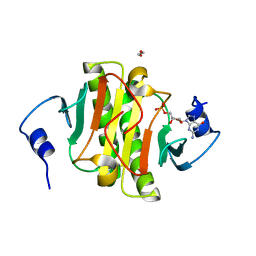
5KM1
 
 | |
5KMA
 
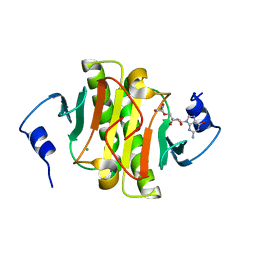 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-ethyl-phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM3
 
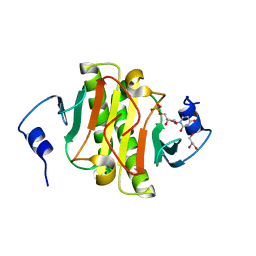 | |
5KMB
 
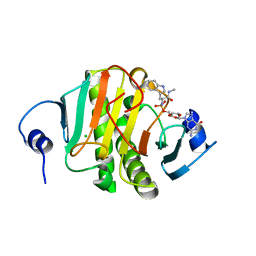 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~ {S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM6
 
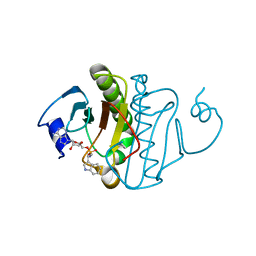 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | Descriptor: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
