4QUO
 
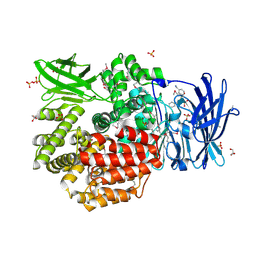 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe(3-CH2NH2) | | Descriptor: | (2S)-2-[3-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
5JW8
 
 | |
4R52
 
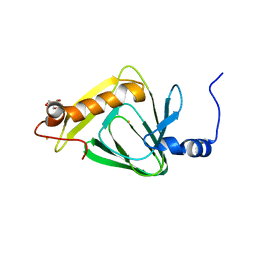 | | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Geng, J, Gumpper, R.H, Huo, L, Liu, A. | | Deposit date: | 2014-08-20 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To be Published
|
|
7TXY
 
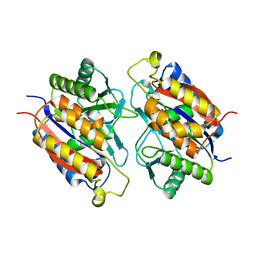 | | Crystal structure of the 2-Aminophenol 1,6-dioxygenase from the ARO bacterial microcompartment of Micromonospora rosaria | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase subunit alpha, 2-aminophenol 1,6-dioxygenase subunit beta, FE (II) ION | | Authors: | Sutter, M, Doron, L, Kerfeld, C.A. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a novel aromatic substrate-processing microcompartment in Actinobacteria.
Mbio, 14, 2023
|
|
7TZ6
 
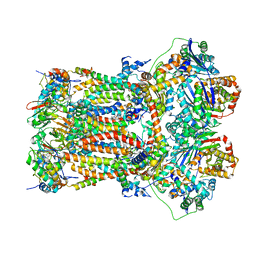 | | Structure of mitochondrial bc1 in complex with ck-2-68 | | Descriptor: | 7-chloranyl-3-methyl-2-[4-[[4-(trifluoromethyloxy)phenyl]methyl]phenyl]-1~{H}-quinolin-4-one, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Huang, R. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of complex III with bound antimalarial agent CK-2-68 provides insights into selective inhibition of Plasmodium cytochrome bc 1 complexes.
J.Biol.Chem., 299, 2023
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
5JYI
 
 | | Trypsin bound with succinic acid at 1.9A | | Descriptor: | CALCIUM ION, Cationic trypsin, SODIUM ION, ... | | Authors: | Manohar, R, Kutumbarao, N.H.V, KarthiK, L, Malathy, P, Velmurugan, D, Gunasekaran, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Trypsin bound with succinic acid at 1.9A
To Be Published
|
|
4X4U
 
 | |
2C8P
 
 | | lysozyme (60sec) and UV laser excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5NWG
 
 | | Crystal structure of TNKS2 in complex with 7-chloro-2-{4-[(2-hydroxyethyl)(methyl)amino]phenyl}-3,4-dihydroquinazolin-4-one | | Descriptor: | 7-chloranyl-2-[4-[2-hydroxyethyl(methyl)amino]phenyl]-3~{H}-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-05-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2-Phenylquinazolinones as dual-activity tankyrase-kinase inhibitors.
Sci Rep, 8, 2018
|
|
7BET
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
5K2B
 
 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
4QT0
 
 | | Crystal structure of human muscle L-lactate dehydrogenase in complex with inhibitor 1, 3-{[3-CARBAMOYL-7-(2,4-DIMETHOXYPYRIMIDIN-5-YL)QUINOLIN-4-YL]AMINO}BENZOIC ACID | | Descriptor: | 3-{[3-carbamoyl-7-(2,4-dimethoxypyrimidin-5-yl)quinolin-4-yl]amino}benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-07-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5K5Y
 
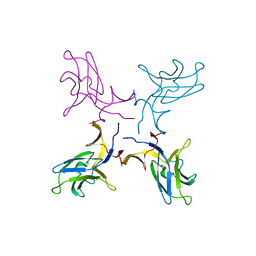 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori (strain 26695) | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Kekez, I, Cendron, L, Stojanovic, M, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Stability of FlgD from the Pathogenic 26695 Strain of Helicobacter pylori
Croatica Chemica Acta, 2016
|
|
4QX2
 
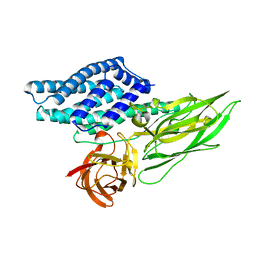 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5K6U
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, IODIDE ION, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
5K92
 
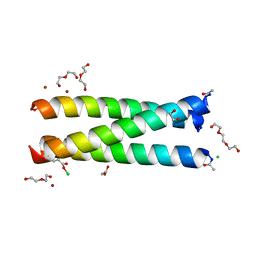 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
4RET
 
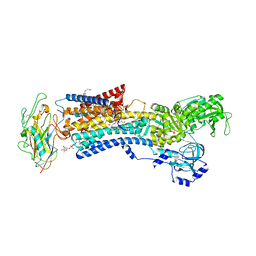 | | Crystal structure of the Na,K-ATPase E2P-digoxin complex with bound magnesium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gregersen, J.L, Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R26
 
 | | Crystal structure of human Fab PGT124, a broadly neutralizing and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGR124-Light Chain, PGT124-Heavy Chain | | Authors: | Garces, F, Kong, L, Wilson, I.A. | | Deposit date: | 2014-08-08 | | Release date: | 2014-10-08 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.4969 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
5K8W
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor U2 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U2
To Be Published
|
|
4X9B
 
 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
5K5G
 
 | | Structure of human islet amyloid polypeptide in complex with an engineered binding protein | | Descriptor: | HI18, Islet amyloid polypeptide | | Authors: | Mirecka, E.A, Feuerstein, S, Gremer, L, Schroeder, G.F, Stoldt, M, Willbold, D, Hoyer, W. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Hairpin of Islet Amyloid Polypeptide Bound to an Aggregation Inhibitor.
Sci Rep, 6, 2016
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
4QT4
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
4WQN
 
 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
