5YJ8
 
 | |
4ZJS
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia Californica (Ac-AChBP) containing the main immunogenic region (MIR) from the human alpha 1 subunit of the muscle nicotinic acetylcholine receptor in complex with anatoxin-A. | | Descriptor: | 1-[(1R,6R)-9-azabicyclo[4.2.1]non-2-en-2-yl]ethanone, Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor,Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J, Park, J.F, Luo, J, Lindsatrom, J, Taylor, P. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2301 Å) | | Cite: | Main immunogenic region structure promotes binding of conformation-dependent myasthenia gravis autoantibodies, nicotinic acetylcholine receptor conformation maturation, and agonist sensitivity.
J. Neurosci., 29, 2009
|
|
5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
3TOA
 
 | | Human MOF crystal structure with active site lysine partially acetylated | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TO6
 
 | |
8EYU
 
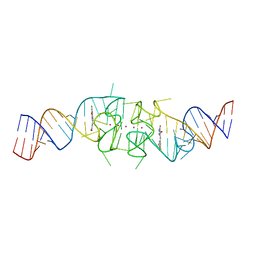 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8EYW
 
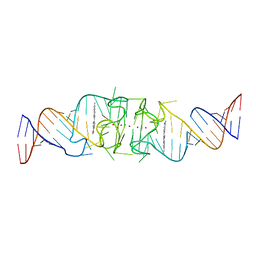 | | Beetroot dimer bound to ThT | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8EYV
 
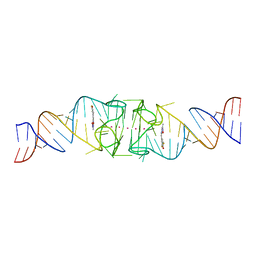 | | Structure of Beetroot dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (45-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8F0N
 
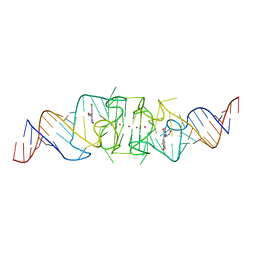 | | Wobble Beetroot (A16U-U38G) dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
5GJQ
 
 | | Structure of the human 26S proteasome bound to USP14-UbAl | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GJR
 
 | | An atomic structure of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
2JRH
 
 | |
5ECE
 
 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3S22
 
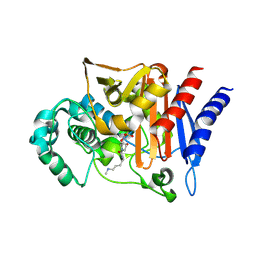 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with an inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, [(2S,3R)-2-formyl-1-{[4-(methylamino)butyl]carbamoyl}pyrrolidin-3-yl]sulfamic acid | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5VXA
 
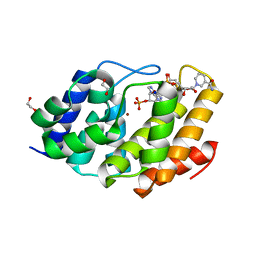 | | Structure of the human Mesh1-NADPH complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
3S1Y
 
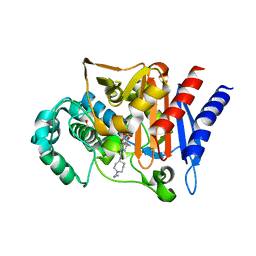 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5EBT
 
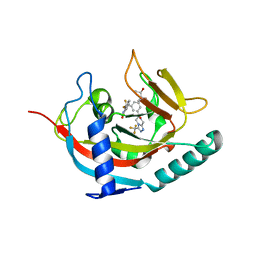 | | Tankyrase 1 with Phthalazinone 2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-[bis(fluoranyl)-[3-[[(6~{S})-6-methyl-3-(trifluoromethyl)-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazin-7-yl]carbonyl]phenyl]methyl]-2~{H}-phthalazin-1-one, Tankyrase-1, ... | | Authors: | Kazmirski, S.L, Johannes, J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4PNZ
 
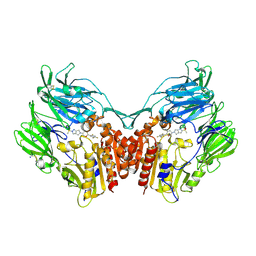 | | Human dipeptidyl peptidase IV/CD26 in complex with the long-acting inhibitor Omarigliptin (MK-3102) | | Descriptor: | (2R,3S,5R)-5-[2-(methylsulfonyl)-2,6-dihydropyrrolo[3,4-c]pyrazol-5(4H)-yl]-2-(2,4,5-trifluorophenyl)tetrahydro-2H-pyran-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Yan, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Omarigliptin (MK-3102): A Novel Long-Acting DPP-4 Inhibitor for Once-Weekly Treatment of Type 2 Diabetes.
J.Med.Chem., 57, 2014
|
|
8QHM
 
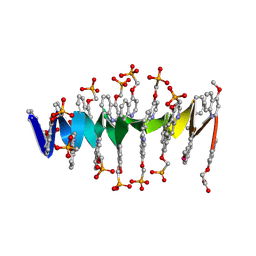 | | DNA mimic Foldamer with sticky ends | | Descriptor: | DNA mimic Foldamer | | Authors: | Deepak, D, Loos, M, Huc, I. | | Deposit date: | 2023-09-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhancing the Features of DNA Mimic Foldamers for Structural Investigations.
Chemistry, 30, 2024
|
|
8SZV
 
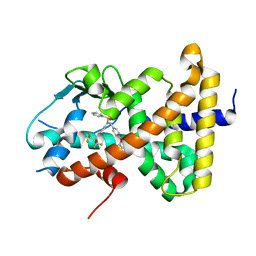 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
4M36
 
 | |
4HT8
 
 | | Crystal structure of E coli Hfq bound to poly(A) A7 | | Descriptor: | Protein hfq, RNA (5'-R(*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Wang, W.W, Wang, L.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA.
Nucleic Acids Res., 41, 2013
|
|
4HT9
 
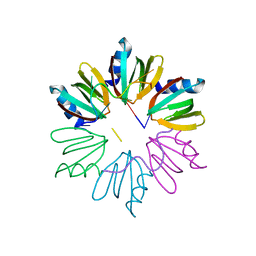 | | Crystal structure of E coli Hfq bound to two RNAs | | Descriptor: | Protein hfq, RNA (5'-R(*AP*AP*AP*AP*AP*AP*A)-3'), RNA (5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3') | | Authors: | Wang, W.W, Wang, L.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA.
Nucleic Acids Res., 41, 2013
|
|
4M37
 
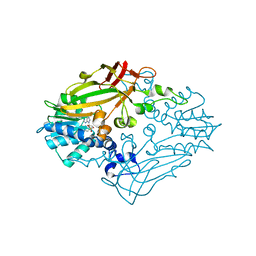 | |
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
