2ID5
 
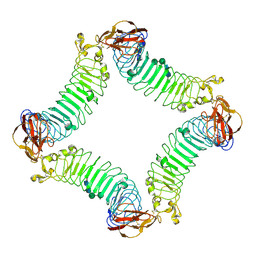 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
8GQU
 
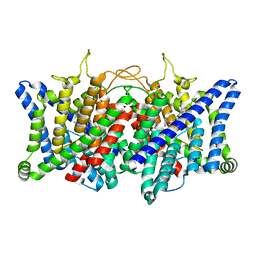 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
3MIL
 
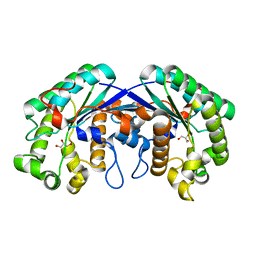 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
7YKS
 
 | |
7YKR
 
 | |
7YPE
 
 | |
7YPF
 
 | |
4PPM
 
 | | Crystal structure of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14 | | Descriptor: | Aminotransferase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lou, X.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
8AXW
 
 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | Authors: | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
4RTA
 
 | |
3JBZ
 
 | |
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
3KZ3
 
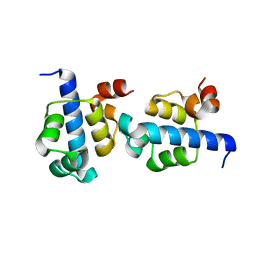 | |
5C8T
 
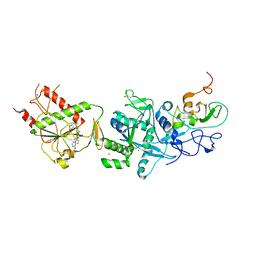 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8U
 
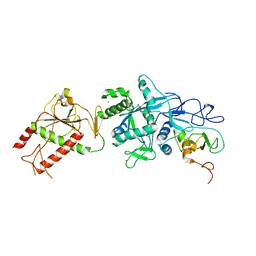 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
5C8S
 
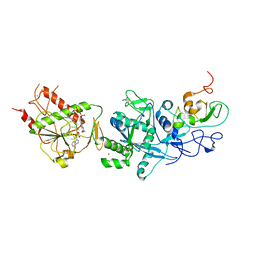 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Guanine-N7 methyltransferase, MAGNESIUM ION, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XRC
 
 | |
5ZYT
 
 | |
5ZUU
 
 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | Descriptor: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Nie, H, Li, S, Patel, D.J. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
