6IGF
 
 | | Crystal structure of Human Papillomavirus type 52 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGE
 
 | | Crystal structure of Human Papillomavirus type 33 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6JGF
 
 | |
6JGV
 
 | |
6JNI
 
 | |
6UZJ
 
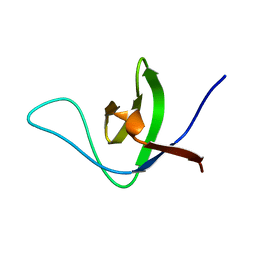 | | NMR structure of the HACS1 SH3 domain | | Descriptor: | SAM domain-containing protein SAMSN-1 | | Authors: | Donaldson, L.W. | | Deposit date: | 2019-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HACS1 signaling adaptor protein recognizes a motif in the paired immunoglobulin receptor B cytoplasmic domain.
Commun Biol, 3, 2020
|
|
8F1D
 
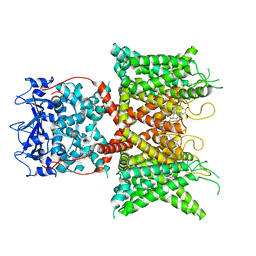 | | Voltage-gated potassium channel Kv3.1 apo | | Descriptor: | CHOLESTEROL, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1C
 
 | | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one, POTASSIUM ION, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4IQK
 
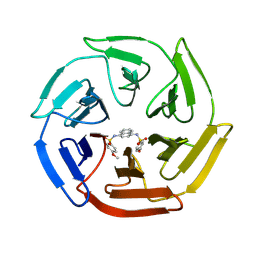 | | Crystal structure of cpd 16 bound to Keap1 Kelch domain | | Descriptor: | Kelch-like ECH-associated protein 1, N,N'-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide) | | Authors: | Silvian, L, Marcotte, D. | | Deposit date: | 2013-01-11 | | Release date: | 2013-05-15 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
7WIY
 
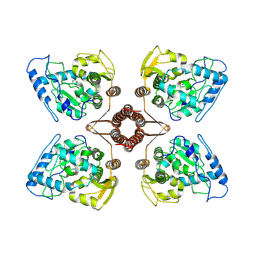 | | Cryo-EM structure of human TPH2 tetramer | | Descriptor: | FE (III) ION, IMIDAZOLE, Tryptophan 5-hydroxylase 2 | | Authors: | Zhu, K.F, Liu, C, Zhang, H.W, Wang, D.P. | | Deposit date: | 2022-01-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM Structure and Activator Screening of Human Tryptophan Hydroxylase 2.
Front Pharmacol, 13, 2022
|
|
8GS4
 
 | | Cryo-EM structure of human Neuroligin 2 | | Descriptor: | Neuroligin-2 | | Authors: | Zhang, H, Zhang, Z, Hou, M. | | Deposit date: | 2022-09-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Expression and structural analysis of human neuroligin 2 and neuroligin 3 implicated in autism spectrum disorders.
Front Endocrinol, 13, 2022
|
|
6IGC
 
 | | Crystal structure of HPV58/33/52 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
3WLD
 
 | | Crystal structure of monomeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
5G3X
 
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6JGX
 
 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JGW
 
 | |
3WLC
 
 | | Crystal structure of dimeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
5YPI
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPK
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPN
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
8SY2
 
 | |
8SY3
 
 | |
8U37
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
