5JFD
 
 | | Thrombin in complex with (S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-D-arginyl)pyrrolidine-2-carboxamide | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
3ZVW
 
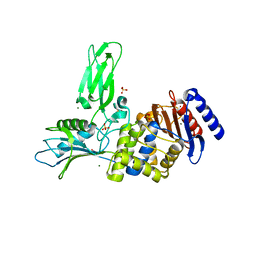 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3-DIMETHYLBUTAN-1-OL, ACETONE, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-07-28 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
3K00
 
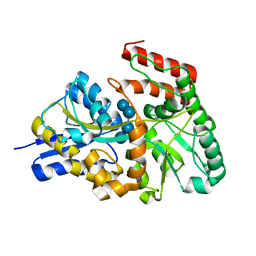 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | Acarbose/maltose binding protein GacH, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
8A39
 
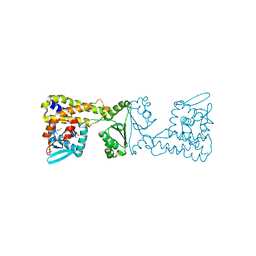 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
7Q4V
 
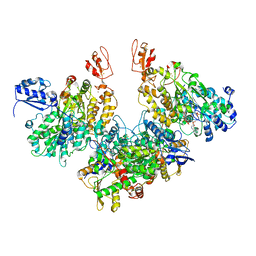 | | Electron bifurcating hydrogenase - HydABC from A. woodii | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Katsyv, A, Kumar, A, Saura, P, Poeverlein, M.C, Freibert, S.A, Stripp, S, Jain, S, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
7Q4W
 
 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
4XUA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
3ZL7
 
 | | BACE2 FYNOMER COMPLEX | | Descriptor: | BETA-SECRETASE 2, FYNOMER 2B-H11 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VIJ
 
 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(1,2,3,4- tetrahydro-1-naphthalenylamino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1S)-1,2,3,4-tetrahydronaphthalen-1-ylamino]propyl}-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Beswick, P, Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Gleave, R, Hawkins, J, Hussain, I, Johnson, C.N, Macpherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Skidmore, J, Soleil, V, Smith, K.J, Stanway, S, Stemp, G, Stuart, A, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-04 | | Release date: | 2008-01-29 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 3: Identification of Hydroxy Ethylamines (Heas) with Nanomolar Potency in Cells.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
8OVE
 
 | |
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
4Y1I
 
 | | Lactococcus lactis yybP-ykoY Mn riboswitch bound to Mn2+ | | Descriptor: | BARIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Lactococcus lactis yybP-ykoY riboswitch, ... | | Authors: | Price, I.R, Ke, A. | | Deposit date: | 2015-02-07 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mn(2+)-Sensing Mechanisms of yybP-ykoY Orphan Riboswitches.
Mol.Cell, 57, 2015
|
|
6RB5
 
 | | Trypanothione reductase in complex with 4-(((3-(8-(2-((1R,2S,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl)ethyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl)propyl)(methyl)amino)methyl)-4-hydroxypiperidine-1-carboximidamide | | Descriptor: | 4-(((3-(8-(2-((1R,2S,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl)ethyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl)propyl)(methyl)amino)methyl)-4-hydroxypiperidine-1-carboximidamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ilari, A, Fiorillo, A, Battista, T, Mocci, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Spiro-containing derivatives show antiparasitic activity against Trypanosoma brucei through inhibition of the trypanothione reductase enzyme.
Plos Negl Trop Dis, 14, 2020
|
|
6R2X
 
 | | NMR structure of Chromogranin A (F39-D63) | | Descriptor: | Chromogranin-A | | Authors: | Nardelli, F, Quilici, G, Ghitti, M, Curnis, F, Gori, A, Berardi, A, Corti, A, Musco, G. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A stapled chromogranin A-derived peptide is a potent dual ligand for integrins alpha v beta 6 and alpha v beta 8.
Chem.Commun.(Camb.), 55, 2019
|
|
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6ZSL
 
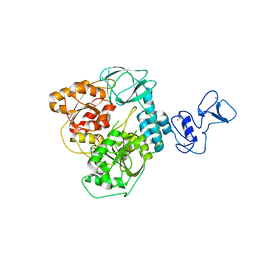 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
3ZKI
 
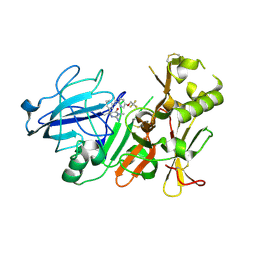 | | BACE2 MUTANT STRUCTURE WITH LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6I8X
 
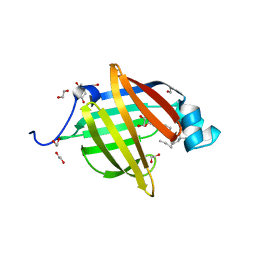 | | As-p18, an extracellular fatty acid binding protein | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein homolog, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Gabrielsen, M, Riboldi-Tunnicliffe, A, Ibanez-Shimabukuro, M, Smith, B.O. | | Deposit date: | 2018-11-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
8I09
 
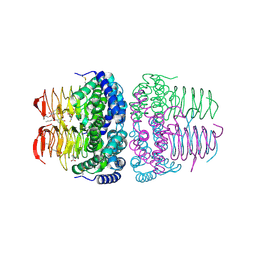 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | Descriptor: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I06
 
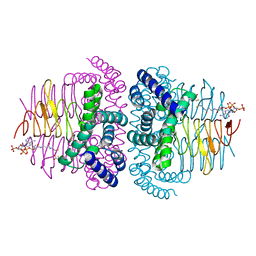 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | Descriptor: | COENZYME A, CYSTEINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
6AZ3
 
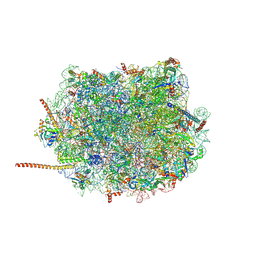 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
6AYB
 
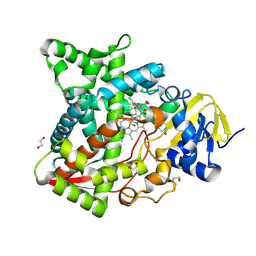 | | Naegleria fowleri CYP51-ketoconazole complex | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, CALCIUM ION, ... | | Authors: | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
8I04
 
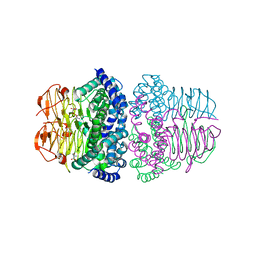 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
5MRT
 
 | | Crystal structure of L5 protease Lysobacter sp. XL1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L5 protease Lysobacter sp. XL1
To Be Published
|
|
