7XLB
 
 | |
4L5B
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the DI-43 inhibitor | | Descriptor: | 1-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
6W2D
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
5HXF
 
 | |
5HXG
 
 | | STM1697-FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Uncharacterized protein STM1697 | | Authors: | Li, B, Yuan, Z, Qin, L, Gu, L. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of STM1697-FlhD complex
To Be Published
|
|
5CZN
 
 | | 2009 H1N1 PA endonuclease mutant E119D | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DBS
 
 | | 2009 H1N1 PA endonuclease mutant E119D in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-21 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5DES
 
 | | 2009 H1N1 PA endonuclease domain | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7YOJ
 
 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
6WWB
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the compound 3b | | Descriptor: | 1,2-ETHANEDIOL, 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-((1-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)acetamido)butyl)-1H-1,2,3-triazol-4-yl)methyl)acetamide, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Phenyl-Glutarimides: Alternative Cereblon Binders for the Design of PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3JSF
 
 | |
9L9X
 
 | | Structure of SPARTA in complex with guide DNA and a 20nt target DNA | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*AP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Hu, R, Guo, C, Liu, X, Chen, J, Liu, L. | | Deposit date: | 2024-12-31 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structural basis of ssDNA-guided NADase activation of prokaryotic SPARTA system.
Nucleic Acids Res., 53, 2025
|
|
9L9W
 
 | | Structure of SPARTA in complex with guide DNA and a 19nt target DNA | | Descriptor: | DNA (5'-D(*T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*AP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Hu, R, Guo, C, Liu, X, Chen, J, Liu, L. | | Deposit date: | 2024-12-31 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (5.87 Å) | | Cite: | Structural basis of ssDNA-guided NADase activation of prokaryotic SPARTA system.
Nucleic Acids Res., 53, 2025
|
|
5L08
 
 | | Cryo-EM structure of Casp-8 tDED filament | | Descriptor: | Caspase-8 | | Authors: | Fu, T.M, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol.Cell, 64, 2016
|
|
6AJ2
 
 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ0
 
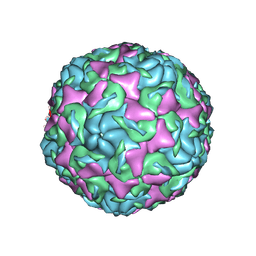 | | The structure of Enterovirus D68 mature virion | | Descriptor: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-25 | | Release date: | 2018-11-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ9
 
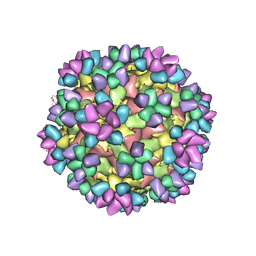 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
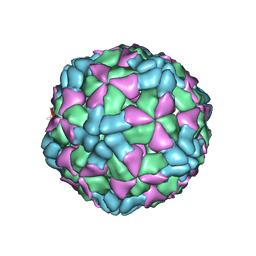 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ7
 
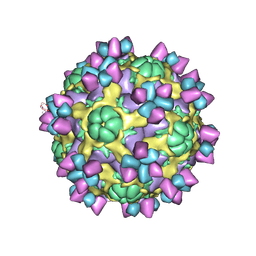 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
7C83
 
 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
6IJI
 
 | | Crystal structure of PDE10 in complex with inhibitor 2b | | Descriptor: | 2-{2-[5-methyl-1-(pyridin-4-yl)-1H-benzimidazol-2-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
6IJH
 
 | | Crystal structure of PDE10 in complex with inhibitor AF-399/14387019 | | Descriptor: | 2-[2-(4-phenyl-5-sulfanylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
