3UCE
 
 | | Crystal structure of a small-chain dehydrogenase in complex with NADPH | | Descriptor: | Dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Buysschaert, G, Verstraete, K, Savvides, S, Vergauwen, B. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference.
Febs J., 280, 2013
|
|
3UCF
 
 | |
1GR5
 
 | | Solution Structure of apo GroEL by Cryo-Electron microscopy | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1GRU
 
 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | Descriptor: | GROEL, GROES | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-16 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
2H50
 
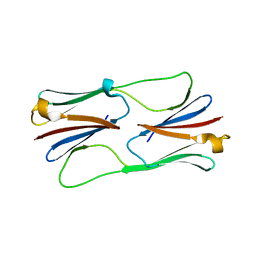 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2H53
 
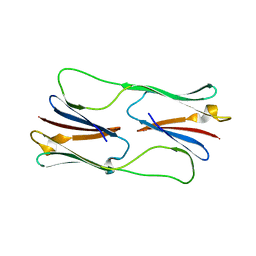 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
5VFC
 
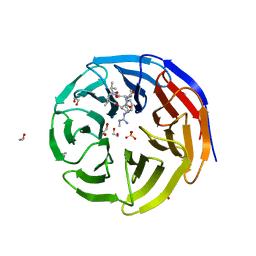 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
5DB2
 
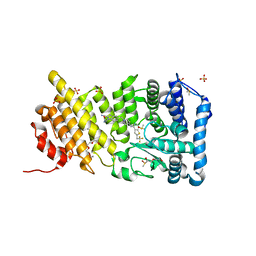 | | Menin in complex with MI-389 | | Descriptor: | 2-{2-cyano-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indol-1-yl}aceta mide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DB1
 
 | | Menin in complex with MI-336 | | Descriptor: | 6-methoxy-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
6WZW
 
 | | Ash1L SET domain in complex with AS-85 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase ASH1L, N-{[3-(3-carbamothioylphenyl)-1-{1-[(trifluoromethyl)sulfonyl]piperidin-4-yl}-1H-indol-6-yl]methyl}azetidine-3-carboxamide, ... | | Authors: | Li, H, Deng, J, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
6X0P
 
 | | Ash1L SET domain Q2265A mutant in complex with AS-5 | | Descriptor: | 3-[6-(aminomethyl)-1-(2-hydroxyethyl)-1H-indol-3-yl]benzene-1-carbothioamide, Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ... | | Authors: | Rogawski, D.S, Li, H, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-17 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
4X5Z
 
 | | menin in complex with MI-136 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
4X5Y
 
 | | Menin in complex with MI-503 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
6O5I
 
 | | Menin in complex with MI-3454 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, SULFATE ION, ... | | Authors: | Linhares, B.M, Klossowski, S, Cierpicki, T, Grembecka, J. | | Deposit date: | 2019-03-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24025619 Å) | | Cite: | Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia.
J.Clin.Invest., 130, 2020
|
|
6BXY
 
 | | Menin in complex with MI-1481 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2S)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
5GMV
 
 | | LC3B-FUNDC1 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FUN14 domain-containing protein 1 | | Authors: | Lv, M, Wang, C, Li, F. | | Deposit date: | 2016-07-17 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy
Protein Cell, 8, 2017
|
|
6BY8
 
 | | Menin in complex with MI-1482 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2R)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, T, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
7MSD
 
 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MSB
 
 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
6W7F
 
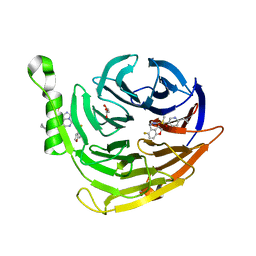 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7G
 
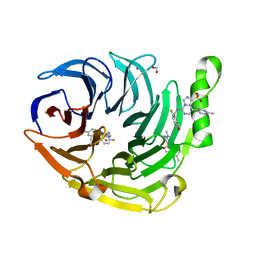 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
