8UAT
 
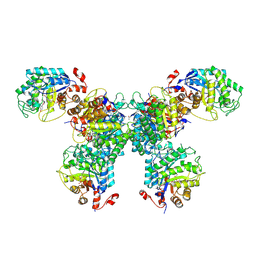 | | Thermus scotoductus SA-01 Ene-reductase Compound 3b Complex | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethyl]-1,4-dihydropyridine-3-carboxamide, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAS
 
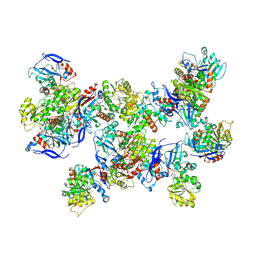 | | Rhodococcus ruber Alcohol Dehydrogenase NADH Biomimetic Complex - Compound 1a | | Descriptor: | 1-[3-[~{tert}-butyl(dimethyl)silyl]oxypropyl]pyridine-3-carboxamide, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAJ
 
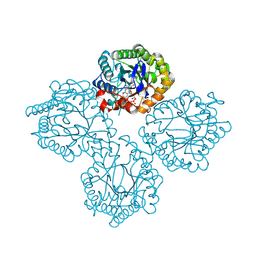 | | Succinate Bound Crystal Structure of Thermus scotoductus SA-01 Ene-reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, SUCCINIC ACID | | Authors: | Wilson, L.A, Guddat, L, Schenk, G, Scott, C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
6C5N
 
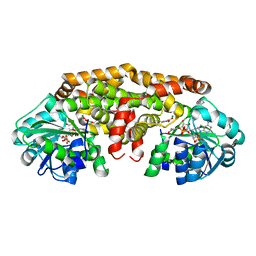 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
6BUL
 
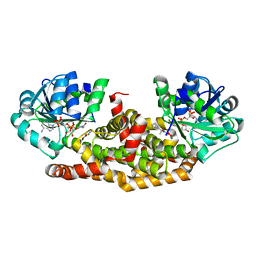 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6C55
 
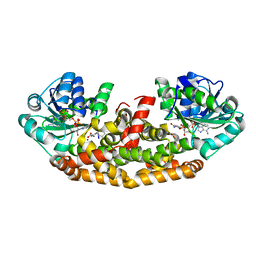 | | Crystal structure of Staphylococcus aureus Ketol-acid Reductosimerrase with hydroxyoxamate inhibitor 3 | | Descriptor: | (cyclohexylamino)(oxo)acetic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schmbri, M, McGeary, R.P. | | Deposit date: | 2018-01-14 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6AQJ
 
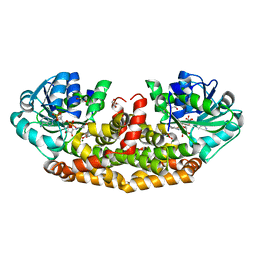 | | Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Gracia, M, Lv, Y, Schembri, M.A, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.373 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
6DR8
 
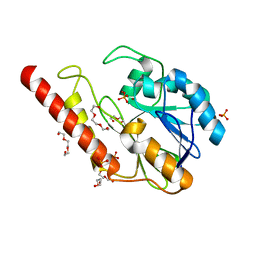 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6DQH
 
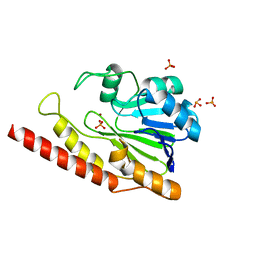 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | Beta-lactamase, PHOSPHATE ION, ZINC ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6DQ2
 
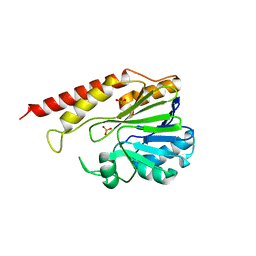 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif mutant S60 | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
7OV3
 
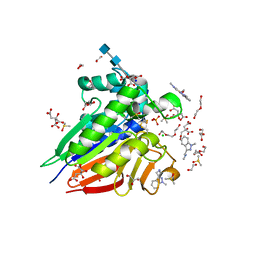 | | Crystal structure of pig purple acid phosphatase in complex with Maybridge fragment CC063346, dimethyl sulfoxide and citrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV2
 
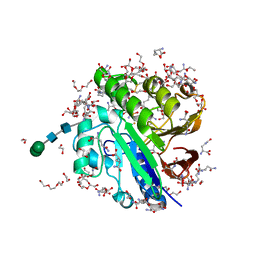 | | Crystal structure of pig purple acid phosphatase in complex with L-glutamine, (poly)ethylene glycol fragments and glycerol | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FE (III) ION, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV8
 
 | | Crystal structure of pig purple acid phosphatase in complex with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and glycerol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
3OOD
 
 | | Structure of OpdA Y257F mutant soaked with diethyl 4-methoxyphenyl phosphate for 20 hours. | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ely, F, Guddat, L.W, Ollis, D.L, Schenk, G. | | Deposit date: | 2010-08-31 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
Biochem.J., 432, 2010
|
|
3OQE
 
 | | Structure of OpdA mutant Y257F | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Ely, F, Guddat, L.W, Ollis, D.L, Schenk, G. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
Biochem.J., 432, 2010
|
|
2QFP
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, FLUORIDE ION, ... | | Authors: | Guddat, L.W, Schenk, G.S, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
5EDJ
 
 | | Crystal structure of the Neisseria meningitidis iron-regulated outer membrane lipoprotein FrpD | | Descriptor: | FrpC operon protein | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
5EDF
 
 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
4YPO
 
 | |
8CY8
 
 | |
8SWM
 
 | |
8SXD
 
 | |
8ET5
 
 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase S653T mutant in complex with amidosulfuron | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of the Commercial Herbicide, Amidosulfuron, in Complex with Arabidopsis thaliana Acetohydroxyacid Synthase.
J.Agric.Food Chem., 71, 2023
|
|
8ET4
 
 | | Crystal structure of wild-type arabidopsis thaliana acetohydroxyacid synthase in complex with amidosulfuron | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Commercial Herbicide, Amidosulfuron, in Complex with Arabidopsis thaliana Acetohydroxyacid Synthase.
J.Agric.Food Chem., 71, 2023
|
|
5VEJ
 
 | |
