7MTZ
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTG
 
 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8TEX
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
8TEY
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
7RKL
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7RKK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (C2 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7SS1
 
 | | The structure of NTMT1 in complex with compound GD433 | | Descriptor: | (1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl {2-[2-(4-fluoro-3-hydroxyphenyl)-1,3-thiazol-4-yl]propan-2-yl}carbamate, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yadav, R, Guangping, D, Deng, Y, Huang, R, Noinaj, N. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a first-in-class small molecule inhibitor for Protein N-terminal methyltransferases 1/2
To Be Published
|
|
7SOK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329 | | Descriptor: | (2S)-2-amino-4-([3-(3-carbamoylphenyl)prop-2-yn-1-yl]{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)butanoic acid, DI(HYDROXYETHYL)ETHER, NNMT protein | | Authors: | Yadav, R, Iyamu, I.D, Huang, R, Noinaj, N. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329
To Be Published
|
|
5VCA
 
 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | Descriptor: | VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5VC7
 
 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
8G2G
 
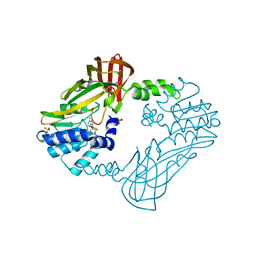 | | Crystal structure of PRMT3 with compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3, SULFATE ION | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2F
 
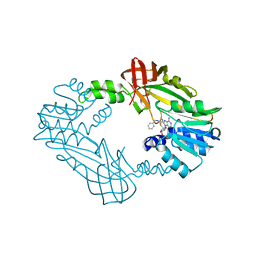 | | Crystal Structure of PRMT3 with Compound II710 | | Descriptor: | 5'-S-[3-(N'-benzylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2H
 
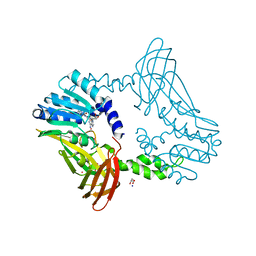 | | Crystal Structure of PRMT4 with Compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2I
 
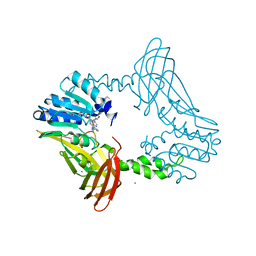 | | Crystal Structure of PRMT4 with Compound YD1290 | | Descriptor: | 5'-([2-(benzylcarbamamido)ethyl]{3-[N'-(3-bromophenyl)carbamimidamido]propyl}amino)-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
6WH8
 
 | | The structure of NTMT1 in complex with compound BM-30 | | Descriptor: | 4HP-PRO-LYS-ARG-NH2, BM-30, N-terminal Xaa-Pro-Lys N-methyltransferase 1, ... | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Selective Peptidomimetic Inhibitors of NTMT1/2: Rational Design, Synthesis, Characterization, and Crystallographic Studies.
J.Med.Chem., 63, 2020
|
|
6PVB
 
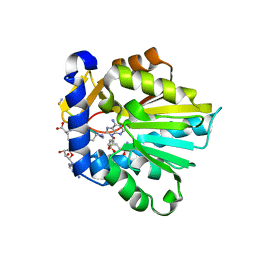 | | The structure of NTMT1 in complex with compound 6 | | Descriptor: | AMINO GROUP-()-(2~{S})-2-azanylpropanal-()-ISOLEUCINE-()-ARGININE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-9-(5-{[(3S)-3-amino-3-carboxypropyl](pentyl)amino}-5-deoxy-beta-L-arabinofuranosyl)-9H-purin-6-amine, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
7N0H
 
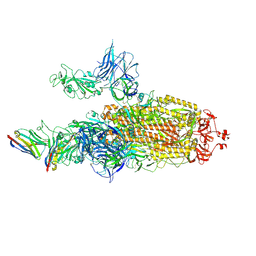 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
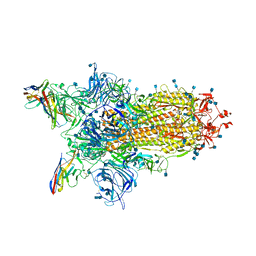 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
6PVA
 
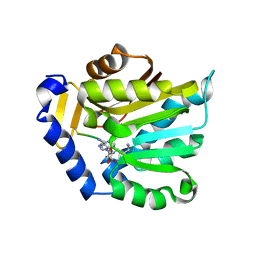 | | The structure of NTMT1 in complex with compound 11 | | Descriptor: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of NTMT1 in complex with compound 11
To Be published
|
|
6UI7
 
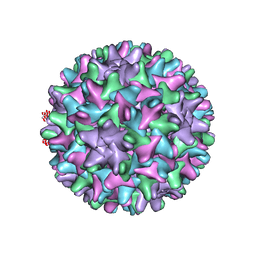 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6UI6
 
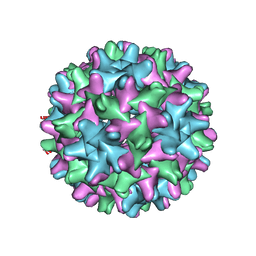 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6WJ7
 
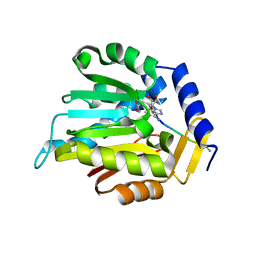 | | The structure of NTMT1 in complex with compound C2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, GLY-PRO-LYS-ARG-ILE-ALA-NH2, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Srinivasan, K, Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
