6GSR
 
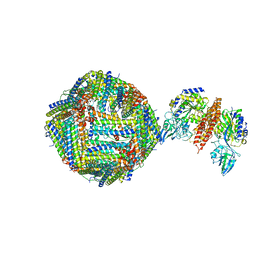 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex at 5.5 Angstrom resolution. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P. | | Deposit date: | 2018-06-15 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
1VYG
 
 | | schistosoma mansoni fatty acid binding protein in complex with arachidonic acid | | Descriptor: | ARACHIDONIC ACID, FATTY ACID BINDING PROTEIN | | Authors: | Angelucci, F, Johnson, K.A, Baiocco, P, Miele, A.E, Bellelli, A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Schistosoma Mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure
Biochemistry, 43, 2004
|
|
1VYF
 
 | | schistosoma mansoni fatty acid binding protein in complex with oleic acid | | Descriptor: | 14 KDA FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Angelucci, F, Johnson, K.A, Baiocco, P, Miele, A.E, Bellelli, A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Schistosoma Mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure
Biochemistry, 43, 2004
|
|
2XWY
 
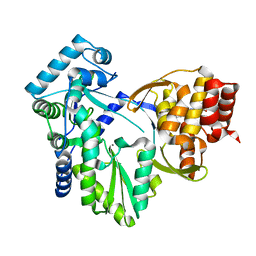 | | Structure of MK-3281, a Potent Non-Nucleoside Finger-Loop Inhibitor, in complex with the Hepatitis C Virus NS5B Polymerase | | Descriptor: | (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl)amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Di Marco, S, Baiocco, P. | | Deposit date: | 2010-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of (7R)-14-Cyclohexyl-7-{[2-(Dimethylamino)Ethyl] (Methyl)Amino}-7,8-Dihydro-6H-Indolo[1,2-E][1,5] Benzoxazocine -11-Carboxylic Acid (Mk-3281), a Potent and Orally Bioavailable Finger-Loop Inhibitor of the Hepatitis C Virus Ns5B Polymerase
J.Med.Chem., 54, 2011
|
|
5YLS
 
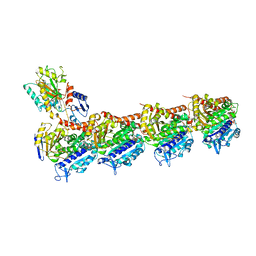 | | Crystal structure of T2R-TTL-Y50 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, E-3-(3-azanyl-4-methoxy-phenyl)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)prop-2-en-1-one, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YLJ
 
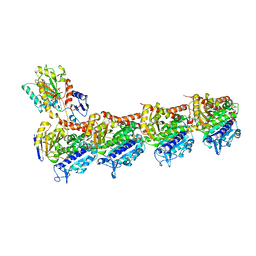 | | Crystal structure of T2R-TTL-Millepachine complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxyphenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
6APD
 
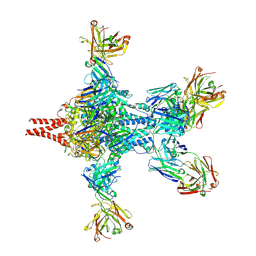 | | Crystal structure of RSV F bound by AM22 and the infant antibody ADI-19425 | | Descriptor: | AM22 Fab Heavy Chain,IGH@ protein, AM22 Fab Light Chain,Uncharacterized protein, Fusion glycoprotein F0,Envelope glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
6APB
 
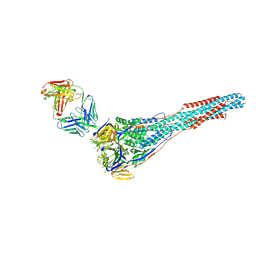 | |
6APC
 
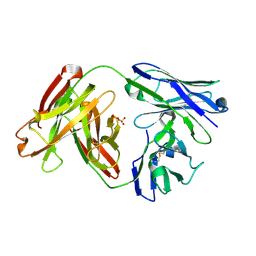 | | Crystal Structure of Infant Antibody ADI-19425 | | Descriptor: | Epididymis luminal protein 214, IGL@ protein, SULFATE ION | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
8WD0
 
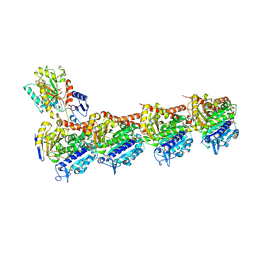 | | Crystal structure of T2R-TTL-Erianin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-[2-(3,4,5-trimethoxyphenyl)ethyl]phenol, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cytotoxic natural compound erianin binds to colchicine site of beta-tubulin and overcomes taxane resistance
Bioorg.Chem., 150, 2024
|
|
8D9C
 
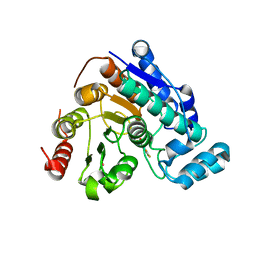 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 10 | | Descriptor: | 2,3,4,5,6-pentafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D99
 
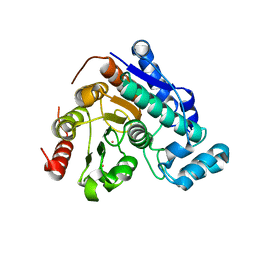 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 7 | | Descriptor: | 2,3,6-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9B
 
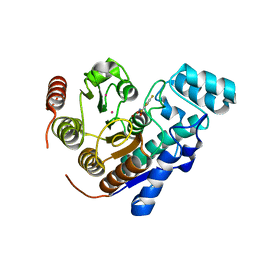 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 9 | | Descriptor: | 2,3,5,6-tetrafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9A
 
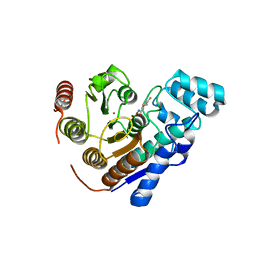 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 8 | | Descriptor: | 2,3,5-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D98
 
 | |
7EXC
 
 | | Crystal structure of T2R-TTL-1129A2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Design and Synthesis of N-Substituted 3-Amino-beta-Carboline Derivatives as Potent alpha beta-Tubulin Degradation Agents
J.Med.Chem., 65, 2022
|
|
8FFJ
 
 | | Structure of Zanidatamab bound to HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, Zanidatamab Heavy Chain A, Zanidatamab Heavy Chain B, ... | | Authors: | Worrall, L.J, Atkinson, C.E, Sanches, M, Dixit, S, Strynadka, N.C.J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Zanidatamab, an Anti-HER2 Biparatopic that Induces Unique Surface HER2 Clusters and Complement-Dependent Cytotoxicity
To be Published
|
|
4FDU
 
 | | Crystal Structure of a Multiple Inositol Polyphosphate Phosphatase | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-HEXASULPHATE, Putative multiple inositol polyphosphate histidine phosphatase 1 | | Authors: | Li, A.W.H, Hemmings, A.M. | | Deposit date: | 2012-05-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut.
Cell Rep, 6, 2014
|
|
4FDT
 
 | | Crystal Structure of a Multiple Inositol Polyphosphate Phosphatase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative multiple inositol polyphosphate histidine phosphatase 1 | | Authors: | Li, A.W.H, Hemmings, A.M. | | Deposit date: | 2012-05-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut.
Cell Rep, 6, 2014
|
|
3S9F
 
 | |
3TUE
 
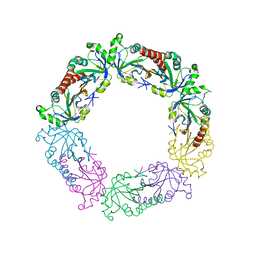 | |
2CK1
 
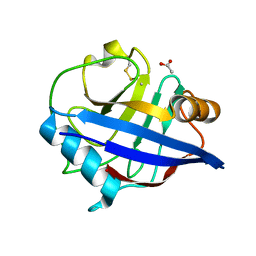 | | The structure of oxidised cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin-A from Schistosoma Mansoni: Evidence for Redox- Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
2CMT
 
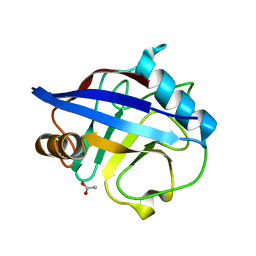 | | The structure of reduced cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin a from Schistosoma Mansoni. Evidence for Redox Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
