6HAR
 
 | | Crystal structure of Mesotrypsin in complex with APPI-M17C/I18F/F34C | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, CALCIUM ION, ... | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J.Biol.Chem., 294, 2019
|
|
1STF
 
 | |
5LLI
 
 | | pVHL:EloB:EloC in complex with VH298 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-cyanocyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent and selective chemical probe of hypoxic signalling downstream of HIF-alpha hydroxylation via VHL inhibition.
Nat Commun, 7, 2016
|
|
6H09
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2018-07-06 | | Release date: | 2018-08-15 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
6J42
 
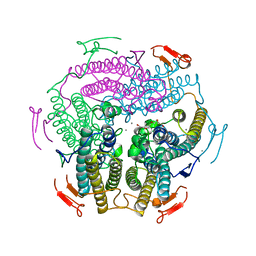 | | Crystal Structure of Wild Type KatB, a manganese catalase from Anabaena | | Descriptor: | Alr3090 protein, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Bihani, S.C, Chakravarty, D, Ballal, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Novel molecular insights into the anti-oxidative stress response and structure-function of a salt-inducible cyanobacterial Mn-catalase.
Plant Cell Environ, 42, 2019
|
|
5V6L
 
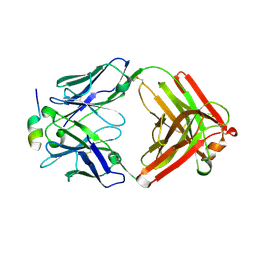 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A37 in complex with V3 peptide JR-FL | | Descriptor: | Envelope glycoprotein, v3 region, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A37, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
5V6M
 
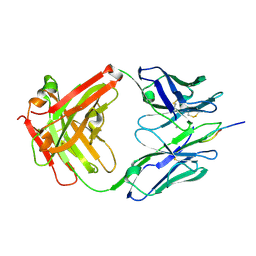 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A3 in complex with V3 peptide ConB | | Descriptor: | CALCIUM ION, Envelope glycoprotein gp120 V3 peptide of Con B sequence, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A3, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
1LDG
 
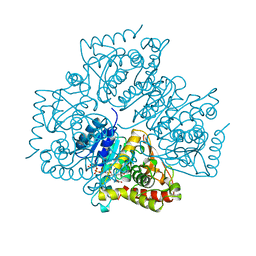 | | PLASMODIUM FALCIPARUM L-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C, Banfield, M, Barker, J, Higham, C, Moreton, K, Turgut-Balik, D, Brady, L, Holbrook, J.J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design.
Nat.Struct.Biol., 3, 1996
|
|
4QN2
 
 | | 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289 | | Descriptor: | ACETATE ION, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QJE
 
 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8PF2
 
 | |
4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OOM
 
 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | Descriptor: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
4Q92
 
 | | 1.90 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-modified Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ES8
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
3IFM
 
 | |
6FDG
 
 | |
6EZ6
 
 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
6EYZ
 
 | | PI3 kinase delta in complex with 4-Fluorophenyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylic acid, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
4L7V
 
 | | Crystal structure of Protein L-isoaspartyl-O-methyltransferase of Vibrio cholerae | | Descriptor: | ACETATE ION, CALCIUM ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Chatterjee, T, Mukherjee, D, Chakrabarti, P. | | Deposit date: | 2013-06-14 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure and activity of protein L-isoaspartyl-O-methyltransferase from Vibrio cholerae, and the effect of AdoHcy binding.
Arch.Biochem.Biophys., 583, 2015
|
|
1FRR
 
 | |
4NEA
 
 | | 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1S6B
 
 | | X-ray Crystal Structure of a Complex Formed Between Two Homologous Isoforms of Phospholipase A2 from Naja naja sagittifera: Principle of Molecular Association and Inactivation | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Jabeen, T, Sharma, S, Singh, R.K, Kaur, P, Singh, T.P. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a calcium-induced dimer of two isoforms of cobra phospholipase A2 at 1.6 A resolution.
Proteins, 59, 2005
|
|
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
