6L7P
 
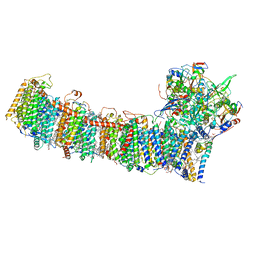 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
4RBX
 
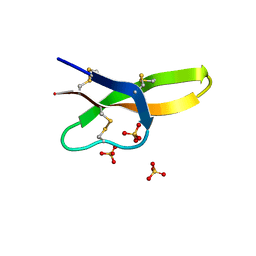 | |
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4RBW
 
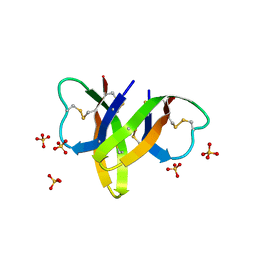 | | Crystal structure of human alpha-defensin 5, HD5 (Thr7Arg Glu21Arg mutant) | | Descriptor: | CHLORIDE ION, Defensin-5, SULFATE ION | | Authors: | Pazgier, M, Gohain, N, Tolbert, W.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a potent antibiotic peptide based on the active region of human defensin 5.
J.Med.Chem., 58, 2015
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
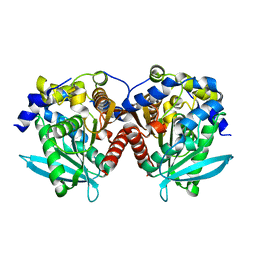
6KMO
 
 | |
5VXA
 
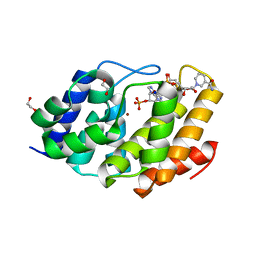 | | Structure of the human Mesh1-NADPH complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
6BAW
 
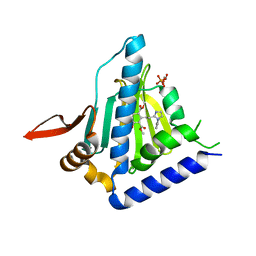 | | Structure of GRP94 with a selective resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmin, PHOSPHATE ION, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
6C91
 
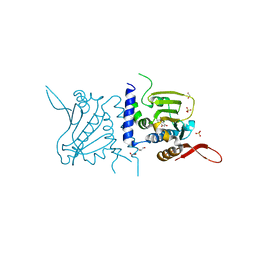 | | Structure of GRP94 with a resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 5-[2-(1-benzyl-1H-imidazol-2-yl)ethyl]-4,6-dichlorobenzene-1,3-diol, Endoplasmin, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WFK
 
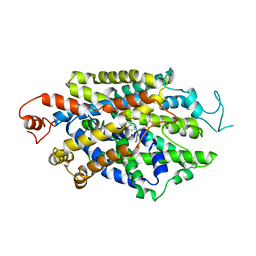 | |
8WFI
 
 | |
8WFL
 
 | |
8WFJ
 
 | |
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
6CEO
 
 | |
5T5G
 
 | | human SETD8 in complex with MS2177 | | Descriptor: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
3H2L
 
 | | Crystal structure of HCV NS5B polymerase in complex with a novel bicyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(4aR,7aS)-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,4a,5,6,7,7a-hexahydro-1H-cyclopenta[b]pyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, NS5B polymerase | | Authors: | Han, Q, Showalter, R.E, Zhou, Q, Kissinger, C.R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of tricyclic 5,6-dihydro-1H-pyridin-2-ones as novel, potent, and orally bioavailable inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5TH7
 
 | | Complex of SETD8 with MS453 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
6KAG
 
 | | Crystal structure of the SMARCB1/SMARCC2 subcomplex | | Descriptor: | SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Chen, G, Zhou, H, Giancotti, F.G, Long, J. | | Deposit date: | 2019-06-22 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | A heterotrimeric SMARCB1-SMARCC2 subcomplex is required for the assembly and tumor suppression function of the BAF chromatin-remodeling complex.
Cell Discov, 6, 2020
|
|
8X5F
 
 | | human XPR1 in complex with InsP6 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
8X5B
 
 | | Cryo-EM structures of human XPR1 in closed states | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, PHOSPHATE ION, ... | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
8X5E
 
 | | Cryo-EM structure of human XPR1 in open state | | Descriptor: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
4MK7
 
 | |
