6K4C
 
 | | Ancestral luciferase AncLamp in complex with DLSA | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, Ancestral luciferase AncLamp, MAGNESIUM ION | | Authors: | Oba, Y, Konishi, K, Yano, D, Kato, D, Shirai, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resurrecting the ancient glow of the fireflies.
Sci Adv, 6, 2020
|
|
5X66
 
 | | Human thymidylate synthase in complex with dUMP and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X5A
 
 | | Human thymidylate synthase bound with phosphate ion | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5J37
 
 | | Crystal structure of 60-mer BFDV Capsid Protein in complex with single stranded DNA | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION, single stranded DNA | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
1K0T
 
 | | NMR SOLUTION STRUCTURE OF UNBOUND, OXIDIZED PHOTOSYSTEM I SUBUNIT PSAC, CONTAINING [4FE-4S] CLUSTERS FA AND FB | | Descriptor: | IRON/SULFUR CLUSTER, PSAC SUBUNIT OF PHOTOSYSTEM I | | Authors: | Antonkine, M.L, Liu, G, Bentrop, D, Bryant, D.A, Bertini, I, Luchinat, C, Golbeck, J.H, Stehlik, D. | | Deposit date: | 2001-09-20 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the unbound, oxidized Photosystem I subunit PsaC, containing [4Fe-4S] clusters F(A) and F(B): a conformational change occurs upon binding to photosystem I.
J.Biol.Inorg.Chem., 7, 2002
|
|
1K2X
 
 | |
1JRK
 
 | | Crystal Structure of a Nudix Protein from Pyrobaculum aerophilum Reveals a Dimer with Intertwined Beta Sheets | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nudix homolog | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5JM4
 
 | | Crystal structure of 14-3-3zeta in complex with a cyclic peptide involving an adamantyl and a dicarboxy side chain | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GLN-GLY-MKD-ANG-ASP-MKD-LEU-ASP-LEU-ALA-CLU | | Authors: | Bier, D, Krueger, D, Glas, A, Wallraven, K, Ottmann, C, Hennig, S, Grossmann, T. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions.
J. Med. Chem., 60, 2017
|
|
1J97
 
 | | Phospho-Aspartyl Intermediate Analogue of Phosphoserine phosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Cho, H, Wang, W, Kim, R, Yokota, H, Damo, S, Kim, S.-H, Wemmer, D, Kustu, S, Yan, D, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6HBH
 
 | | Echovirus 18 A-particle | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
6HUJ
 
 | | CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with picrotoxin, GABA and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
1IH8
 
 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6HLF
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant - K32A | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Crystal Contact Engineering of Lactobacillus brevis Alcohol Dehydrogenase To Promote Technical Protein Crystallization
Cryst.Growth Des., 2019
|
|
5X2W
 
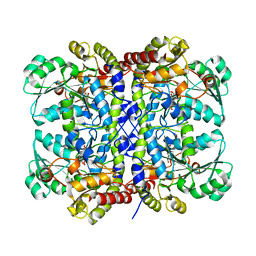 | |
5XG5
 
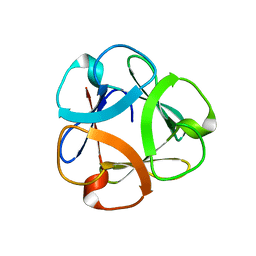 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
1J8Q
 
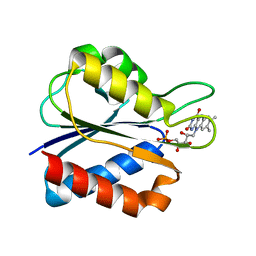 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris Wild-type at 1.35 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-22 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IG6
 
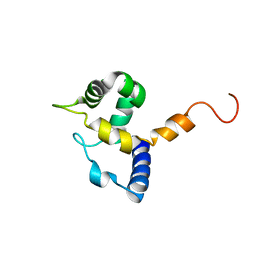 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | Descriptor: | MODULATOR RECOGNITION FACTOR 2 | | Authors: | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
4OW3
 
 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
1EPG
 
 | |
1K76
 
 | | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J, Volk, D, Luxon, B, Gorenstein, D, Hilser, V. | | Deposit date: | 2001-10-18 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
4O9R
 
 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
6K7U
 
 | | Crystal structure of beta-2 microglobulin (beta2m) of Bat (Pteropus Alecto) | | Descriptor: | Bat beta-2-microglobulin | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
1K8X
 
 | | Crystal Structure Of AlphaT183V Mutant Of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-10-26 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of AlphaThr183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1K3B
 
 | | Crystal Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Turk, D, Janjic, V, Stern, I, Podobnik, M, Lamba, D, Dahl, S.W, Lauritzen, C, Pedersen, J, Turk, V, Turk, B. | | Deposit date: | 2001-10-02 | | Release date: | 2002-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases.
EMBO J., 20, 2001
|
|
1RNO
 
 | | RIBONUCLEASE A CRYSTALLIZED FROM 80% AMMONIUM SULPHATE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | Deposit date: | 1995-11-08 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
