6KYR
 
 | |
4ZRI
 
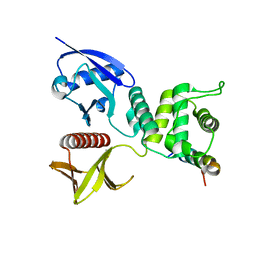 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
5HMO
 
 | |
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
5ZNR
 
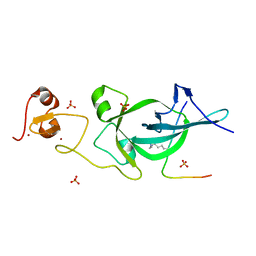 | | Crystal structure of PtSHL in complex with an H3K27me3 peptide | | Descriptor: | 17-mer peptide from Histone H3.2, SHORT LIFE family protein, SULFATE ION, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
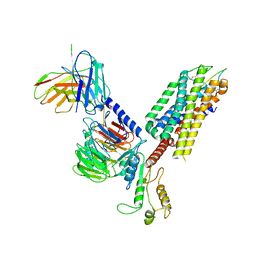 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
5ZWZ
 
 | |
7F8X
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
5ZWX
 
 | |
7CKG
 
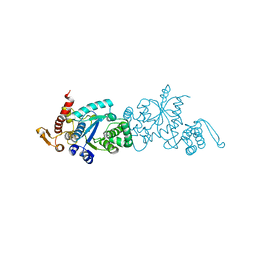 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKH
 
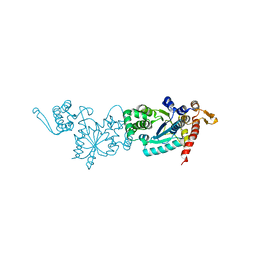 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
2O7N
 
 | | CD11A (LFA1) I-domain complexed with 7A-[(4-cyanophenyl)methyl]-6-(3,5-dichlorophenyl)-5-oxo-2,3,5,7A-tetrahydro-1H-pyrrolo[1,2-A]pyrrole-7-carbonitrile | | Descriptor: | 7A-[(4-cyanophenyl)methyl]-6-(3,5-dichlorophenyl)-5-oxo-2,3,5,7A-tetrahydro-1H-pyrrolo[1,2-A]pyrrole-7-carbonitrile, Integrin alpha-L | | Authors: | Sheriff, S. | | Deposit date: | 2006-12-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of LFA-1 antagonists based on a 2,3-dihydro-1H-pyrrolizin-5(7aH)-one scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6IHB
 
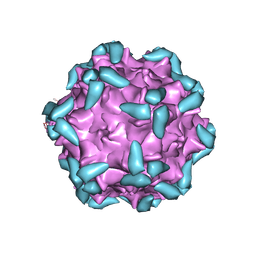 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
6IH9
 
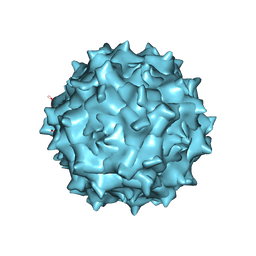 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
4RHD
 
 | | DNA Duplex with Novel ZP Base Pair | | Descriptor: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | Authors: | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
4P7I
 
 | | Crystal structure of the Merlin FERM/DCAF1 complex | | Descriptor: | GLYCEROL, Merlin, Protein VPRBP | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the binding of Merlin FERM domain to the E3 ubiquitin ligase substrate adaptor DCAF1.
J.Biol.Chem., 289, 2014
|
|
2GM1
 
 | | Crystal structure of the mitotic kinesin eg5 in complex with mg-adp and n-(3-aminopropyl)-n-((3-benzyl-5-chloro-4-oxo-3,4-dihydropyrrolo[2,1-f][1,2,4]triazin-2-yl)(cyclopropyl)methyl)-4-methylbenzamide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-RELATED MOTOR PROTEIN EG5, MAGNESIUM ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and SAR of pyrrolotriazine-4-one based Eg5 inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8C5V
 
 | |
3R24
 
 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
6IH0
 
 | | Aquifex aeolicus LpxC complex with ACHN-975 | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Li, D.Y, Fan, S, Jin, Y.Y, Zhang, C, Lv, G.X, Wu, G.T, Yang, Z.Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Complex Structure of Protein AaLpxC from Aquifex aeolicus with ACHN-975 Molecule Suggests an Inhibitory Mechanism at Atomic-Level against Gram-Negative Bacteria
Molecules, 26, 2021
|
|
7RCB
 
 | | Crystal Structure of a PMS2 VUS | | Descriptor: | Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
