4QQJ
 
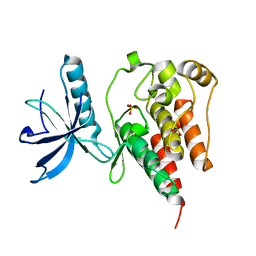 | |
4QQT
 
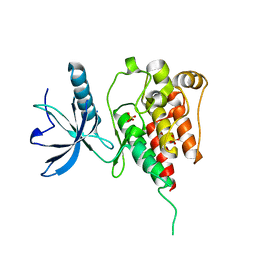 | |
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
6ATE
 
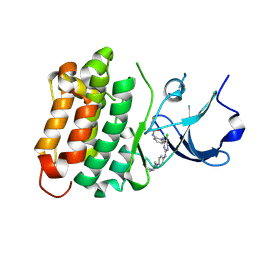 | | SRC kinase bound to covalent inhibitor | | Descriptor: | N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6BEA
 
 | |
5Y2T
 
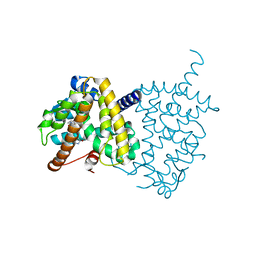 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5Y2O
 
 | | Structure of PPARgamma ligand binding domain-pioglitazone complex | | Descriptor: | (5S)-5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
7D6H
 
 | |
7E35
 
 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
7POM
 
 | |
7E4Z
 
 | | Crystal structure of tubulin in complex with Maytansinol | | Descriptor: | (1R,2S,3S,5S,6S,16E,18E,20R,21S)-11-chloro-6,21-dihydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4Q
 
 | | Crystal structure of tubulin in complex with L-DM1-SMe | | Descriptor: | (1S,2R,3R,5R,6R,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methyldisulfanyl)propanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4R
 
 | | Crystal structure of tubulin in complex with D-DM1-SMe | | Descriptor: | (10E,12E)-86-chloro-14-hydroxy-85,14-dimethoxy-33,2,7,10-tetramethyl-12,6-dioxo-7-aza-1(6,4)-oxazinana-3(2,3)-oxirana-8(1,3)-benzenacyclotetradecaphane-10,12-dien-4-yl N-methyl-N-(3-(methylsulfinothioyl)propanoyl)-D-alaninate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7PUU
 
 | | Crystal structure of carbonic anhydrase XII with methyl 4-chloro-2-cyclohexylsulfanyl-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUW
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUV
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0C
 
 | | Mimic carbonic anhydrase IX in complex with Methyl 2-chloro-4-(cyclohexylsulfanyl)-5-sulfamoylbenzoate | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0D
 
 | | Human carbonic anhydrase I in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0E
 
 | | Human Carbonic Anhydrase II in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
3ZTX
 
 | | Aurora kinase selective inhibitors identified using a Taxol-induced checkpoint sensitivity screen. | | Descriptor: | 2-((4-(4-HYDROXYPIPERIDIN-1-YL)PHENYL)AMINO)-5,11-DIMETHYL-5H-BENZO[E]PYRIMIDO [5,4-B][1,4]DIAZEPIN-6(11H)-ONE, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Kwiatkowski, N, Villa, F, Musacchio, A, Gray, N. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Aurora Kinase Inhibitors Identified Using a Taxol- Induced Checkpoint Sensitivity Screen.
Acs Chem.Biol., 7, 2012
|
|
5VAM
 
 | | BRAF in Complex with RAF709 | | Descriptor: | N-{2-methyl-5'-(morpholin-4-yl)-6'-[(oxan-4-yl)oxy][3,3'-bipyridin]-5-yl}-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
5VAL
 
 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
1D2V
 
 | | CRYSTAL STRUCTURE OF BROMIDE-BOUND HUMAN MYELOPEROXIDASE ISOFORM C AT PH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BROMIDE ION, ... | | Authors: | Fiedler, T.J, Davey, C.A, Fenna, R.E. | | Deposit date: | 1999-09-28 | | Release date: | 2000-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure and characterization of halide-binding sites of human myeloperoxidase at 1.8 A resolution.
J.Biol.Chem., 275, 2000
|
|
1D5L
 
 | | CRYSTAL STRUCTURE OF CYANIDE-BOUND HUMAN MYELOPEROXIDASE ISOFORM C AT PH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Fiedler, T.J, Davey, C.A, Fenna, R.E. | | Deposit date: | 1999-10-07 | | Release date: | 2001-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 A resolution.
Biochemistry, 40, 2001
|
|
