4NTI
 
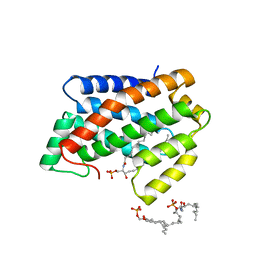 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTG
 
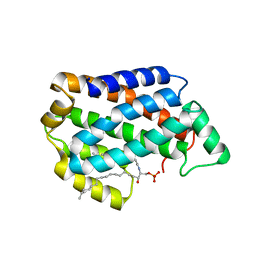 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
1RA4
 
 | |
7W3T
 
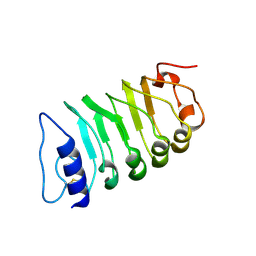 | | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3X
 
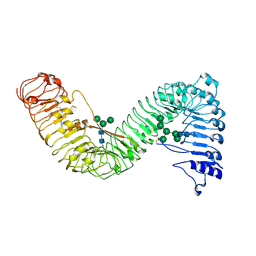 | | Cryo-EM structure of plant receptor like protein RXEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-localized LRR receptor-like protein, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3V
 
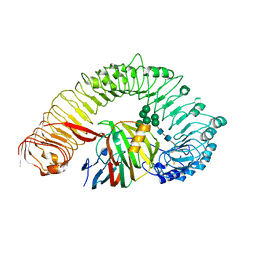 | | Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
6JWE
 
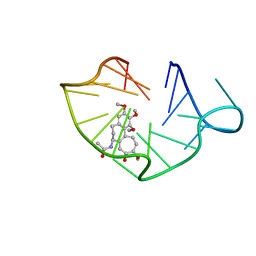 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
7VZN
 
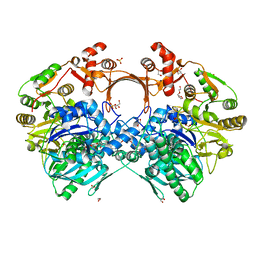 | | The structure of GdmN in complex with carbamoyl adenylate intermediate and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7CFS
 
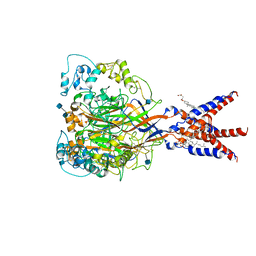 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
6KWD
 
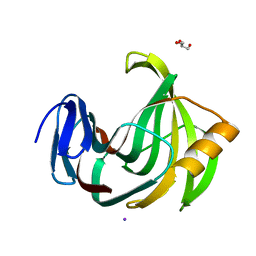 | | Crystal Structure Analysis of Endo-beta-1,4-Xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
7CFT
 
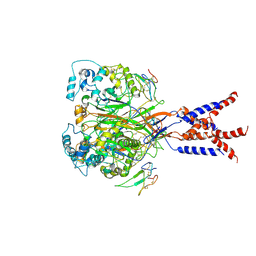 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
5XDM
 
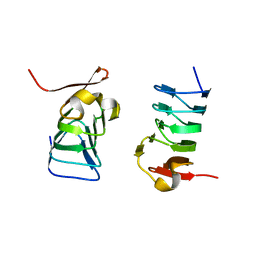 | |
7CBQ
 
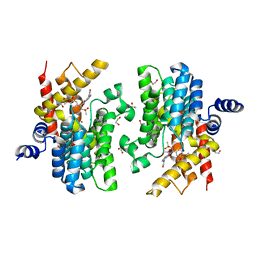 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
5XLP
 
 | |
5XOE
 
 | |
6KW9
 
 | |
6KWF
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6KWG
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
6KWC
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
5XZ9
 
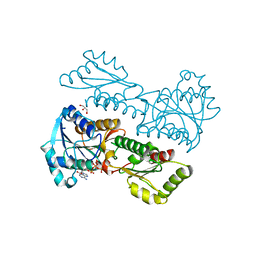 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, GLYCEROL | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-12 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ7
 
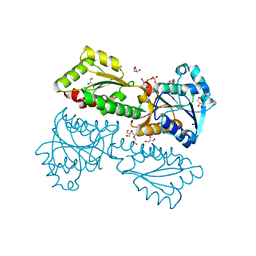 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZA
 
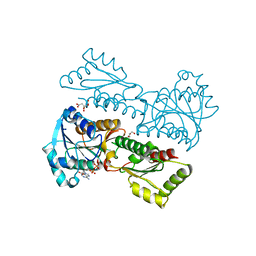 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, CITRATE ANION, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-12 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ8
 
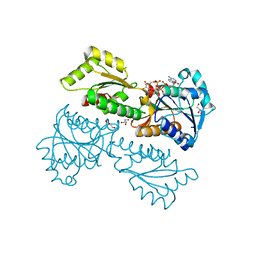 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate (the ATP analog) and fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, GLYCEROL, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
