6KJZ
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 1 | | Descriptor: | 5,9-dihydroxy-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H,6H-pyrano[3,2-b]xanthen-6-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
6B41
 
 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 98, 2024
|
|
3WBM
 
 | | Crystal Structure of protein-RNA complex | | Descriptor: | DNA/RNA-binding protein Alba 1, RNA (25-MER) | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea.
J.Biol.Chem., 289, 2014
|
|
8ID1
 
 | |
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
8IOF
 
 | |
1S13
 
 | | Human Heme Oxygenase Oxidatition of alpha- and gamma-meso-Phenylhemes | | Descriptor: | 2-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-01-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
1T5P
 
 | | Human Heme Oxygenase Oxidation of alpha- and gamma-meso-phenylhemes | | Descriptor: | 12-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
8KAG
 
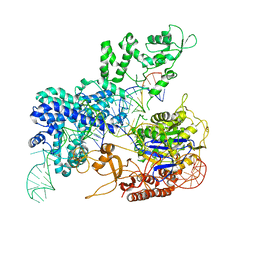 | | Crystal structure of SpyCas9 in complex with sgRNA and target RNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, RNA (5'-R(*CP*CP*AP*CP*UP*UP*CP*AP*AP*UP*UP*AP*GP*AP*AP*CP*AP*CP*GP*GP*AP*CP*C)-3'), RNA (98-MER) | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAK
 
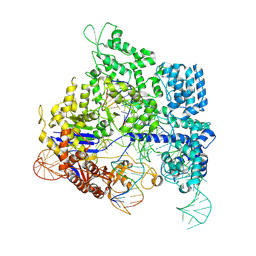 | |
8KAJ
 
 | | Crystal structure of SpyCas9-crRNA-tracrRNA complex bound to 16nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*AP*AP*TP*AP*CP*CP*TP*TP*TP*TP*AP*TP*CP*CP*AP*TP*AP*AP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAH
 
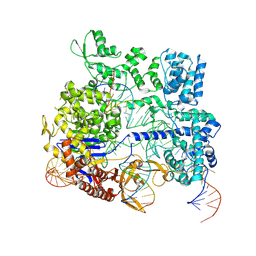 | |
8KAI
 
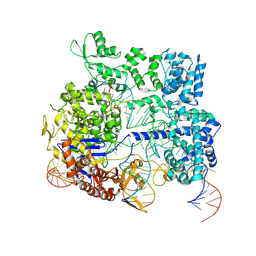 | |
8KAM
 
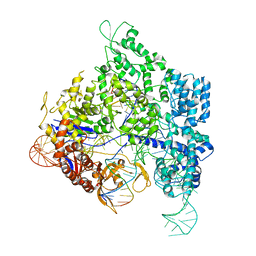 | | Crystal structure of SpyCas9 in complex with sgRNA and 16nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*AP*AP*TP*AP*CP*CP*TP*CP*TP*TP*CP*AP*AP*TP*TP*AP*GP*AP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAL
 
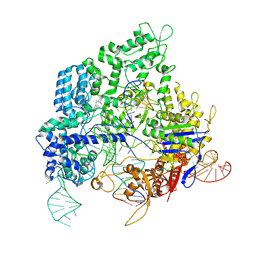 | |
5VFC
 
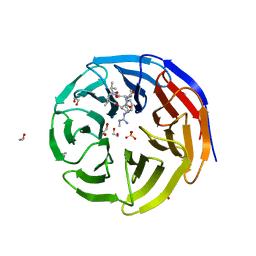 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
7QXP
 
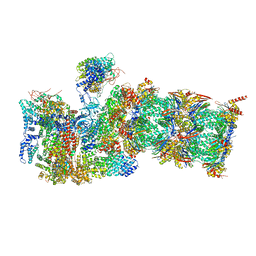 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXX
 
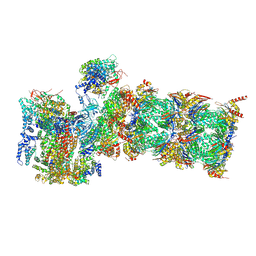 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXU
 
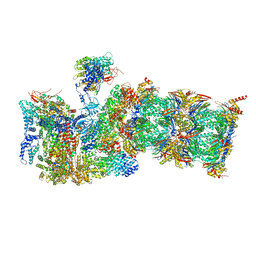 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QY7
 
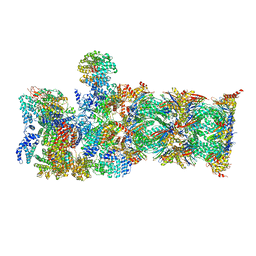 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXW
 
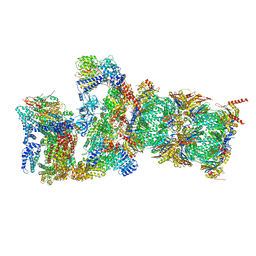 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXN
 
 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
