2ZBJ
 
 | | Crystal structure of Dioclea rostrata lectin | | 分子名称: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION, ... | | 著者 | de Oliveira, T.M, Delatorre, P, da Rocha, B.A.M, de Sousa, E.P, Nascimento, K.S, Bezerra, G.A, Moura, T.R, Benevides, R.G, Bezerra, E.H.S, Moreno, F.B.M.B, Freire, V.N, de Azevedo Jr, W.F, Cavada, B.S. | | 登録日 | 2007-10-22 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of Dioclea rostrata lectin: Insights into understanding the pH-dependent dimer-tetramer equilibrium and the structural basis for carbohydrate recognition in Diocleinae lectins
J.Struct.Biol., 164, 2008
|
|
2EIG
 
 | | Lotus tetragonolobus seed lectin (Isoform) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | 著者 | Moreno, F.B.M.B, Vicoti, M.M, Abrego, J.R.B, de Oliveira, T.M, Bezerra, G.A, Cavada, B.S, Filgueira de Azevedo Jr, W. | | 登録日 | 2007-03-13 | | 公開日 | 2008-03-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a new quaternary association for legume lectins
J.Struct.Biol., 161, 2008
|
|
3AMB
 
 | | Protein kinase A sixfold mutant model of Aurora B with inhibitor VX-680 | | 分子名称: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Pflug, A, de Oliveira, T.M, Bossemeyer, D, Engh, R.A. | | 登録日 | 2010-08-18 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Biochem.J., 440, 2011
|
|
3AMA
 
 | | Protein kinase A sixfold mutant model of Aurora B with inhibitor JNJ-7706621 | | 分子名称: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Pflug, A, de Oliveira, T.M, Bossemeyer, D, Engh, R.A. | | 登録日 | 2010-08-18 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Biochem.J., 440, 2011
|
|
2OW4
 
 | | Crystal structure of a lectin from Canavalia maritima seeds (ConM) in complex with man1-2man-OMe | | 分子名称: | CALCIUM ION, Canavalia maritima lectin, MANGANESE (II) ION, ... | | 著者 | Moreno, F.B.M.B, Bezerra, G.A, de Oliveira, T.M, de Souza, E.M, da Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W.F. | | 登録日 | 2007-02-15 | | 公開日 | 2007-10-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
2OVU
 
 | | Crystal structure of a lectin from Canavalia gladiata (CGL) in complex with man1-2man-OMe | | 分子名称: | CALCIUM ION, Canavalia gladiata lectin, MANGANESE (II) ION, ... | | 著者 | Moreno, F.B.M.B, Bezerra, G.A, de Oliveira, T.M, de Souza, E.P, da Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W.F. | | 登録日 | 2007-02-15 | | 公開日 | 2007-10-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
6ZHA
 
 | | Cryo-EM structure of DNA-PK monomer | | 分子名称: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Varela, P.F, Charbonnier, J.B. | | 登録日 | 2022-03-14 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
6ZFP
 
 | | Cryo-EM structure of DNA-PKcs (State 2) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
 | | Cryo-EM structure of DNA-PK dimer | | 分子名称: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
 | | Cryo-EM structure of DNA-PKcs (State 1) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
 | | Cryo-EM structure of DNA-PKcs (State 3) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH8
 
 | | Cryo-EM structure of DNA-PKcs:DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ZT6
 
 | |
7ZVT
 
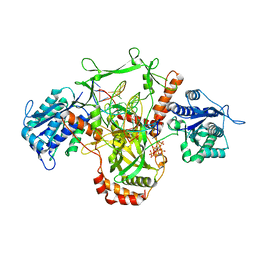 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
8BOT
 
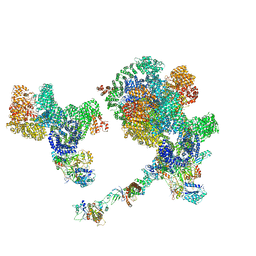 | |
7NFC
 
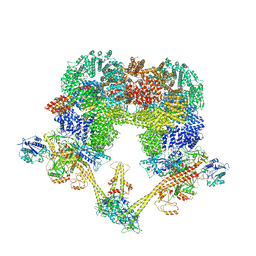 | | Cryo-EM structure of NHEJ super-complex (dimer) | | 分子名称: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-05 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
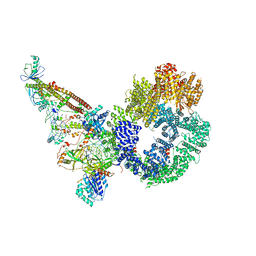 | | Cryo-EM structure of NHEJ super-complex (monomer) | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-06 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
