1INP
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE AT 2.3 ANGSTROMS RESOLUTION | | 分子名称: | INOSITOL POLYPHOSPHATE 1-PHOSPHATASE, MAGNESIUM ION | | 著者 | York, J.D, Ponder, J.W, Chen, Z, Mathews, F.S, Majerus, P.W. | | 登録日 | 1994-10-04 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution.
Biochemistry, 33, 1994
|
|
6X25
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | 分子名称: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | 著者 | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | 登録日 | 2020-05-20 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6WRO
 
 | |
6WRY
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AT 2.5 ANGSTROM RESOLUTION | | 分子名称: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | 著者 | Dollins, D.R, Endo-Streeter, S, York, J.D. | | 登録日 | 2020-04-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6WRR
 
 | |
4XNH
 
 | |
4XPD
 
 | |
6PCK
 
 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 1-IP7 | | 分子名称: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | 著者 | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | 登録日 | 2019-06-17 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Y49
 
 | |
6PCL
 
 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | 分子名称: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | 著者 | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | 登録日 | 2019-06-17 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1I9Y
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN | | 分子名称: | PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | 著者 | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | 登録日 | 2001-03-21 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I9Z
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN IN COMPLEX WITH INOSITOL (1,4)-BISPHOSPHATE AND CALCIUM ION | | 分子名称: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | 著者 | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | 登録日 | 2001-03-21 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
7KIR
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | 分子名称: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | 著者 | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7KIO
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant | | 分子名称: | CALCIUM ION, Inositol polyphosphate 1-phosphatase, SULFATE ION | | 著者 | Xiong, J.-P, Dollins, D.E, Ren, Y, York, J.D. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
4ITL
 
 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | 分子名称: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4ITN
 
 | | Crystal structure of "compact P-loop" LpxK from Aquifex aeolicus in complex with chloride at 2.2 angstrom resolution | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1912 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4ITM
 
 | | Crystal structure of "apo" form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1994 Å) | | 主引用文献 | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4LKV
 
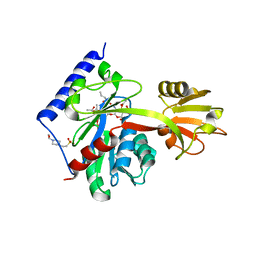 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | 分子名称: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | 登録日 | 2013-07-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5109 Å) | | 主引用文献 | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
4FRF
 
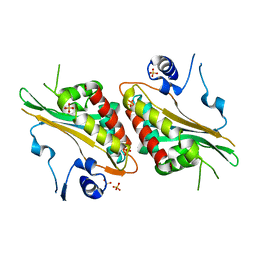 | | Structural Studies and Protein Engineering of Inositol Phosphate Multikinase | | 分子名称: | Inositol polyphosphate multikinase alpha, SULFATE ION | | 著者 | Endo-Streeter, S.T, Tsui, M, Odom, A.R, York, J.D. | | 登録日 | 2012-06-26 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural studies and protein engineering of inositol phosphate multikinase.
J.Biol.Chem., 287, 2012
|
|
4HNY
 
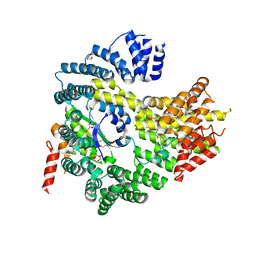 | | Apo N-terminal acetyltransferase complex A | | 分子名称: | GLYCEROL, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | 著者 | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | 登録日 | 2012-10-21 | | 公開日 | 2014-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.249 Å) | | 主引用文献 | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNW
 
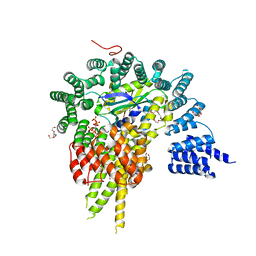 | | The NatA Acetyltransferase Complex Bound To Inositol Hexakisphosphate | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | 著者 | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | 登録日 | 2012-10-21 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNX
 
 | | The NatA Acetyltransferase Complex Bound To ppGpp | | 分子名称: | GUANOSINE-5',3'-TETRAPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | 著者 | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | 登録日 | 2012-10-21 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.339 Å) | | 主引用文献 | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
6D74
 
 | |
5FSA
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from a pathogenic yeast Candida albicans in complex with the antifungal drug posaconazole | | 分子名称: | CYP51 VARIANT1, POSACONAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hargrove, T.Y, Wawrzak, Z, Friggeri, L, Lepesheva, G.I. | | 登録日 | 2016-01-02 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5TZ1
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Candida albicans in complex with the tetrazole-based antifungal drug candidate VT1161 (VT1) | | 分子名称: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | 著者 | Hargrove, T, Wawrzak, Z, Lepesheva, G. | | 登録日 | 2016-11-21 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
