4AM3
 
 | | Crystal structure of C. crescentus PNPase bound to RNA | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-03-07 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AIM
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-10 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AID
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
2J6Y
 
 | | Structural and Functional Characterisation of partner switching regulating the environmental stress response in Bacillus subtilis | | 分子名称: | PHOSPHOSERINE PHOSPHATASE RSBU | | 著者 | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | 登録日 | 2006-10-05 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2J6Z
 
 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | 分子名称: | PHOSPHOSERINE PHOSPHATASE RSBU | | 著者 | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | 登録日 | 2006-10-05 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2J70
 
 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | 分子名称: | PHOSPHOSERINE PHOSPHATASE RSBU | | 著者 | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | 登録日 | 2006-10-05 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
8BOT
 
 | |
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZWA
 
 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-19 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | 分子名称: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8BH3
 
 | |
8BHY
 
 | |
8BHV
 
 | |
6GWK
 
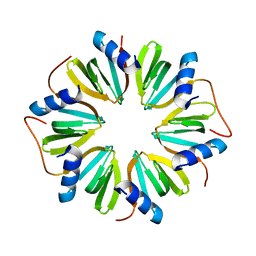 | | The crystal structure of Hfq from Caulobacter crescentus | | 分子名称: | RNA-binding protein Hfq | | 著者 | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | 登録日 | 2018-06-25 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5FT1
 
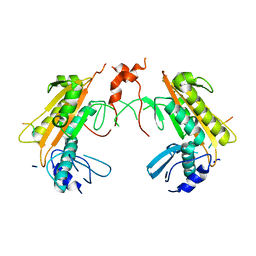 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ bound to RNase E of Pseudomonas aeruginosa | | 分子名称: | GP37, RIBONUCLEASE E | | 著者 | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | 登録日 | 2016-01-08 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
5FT0
 
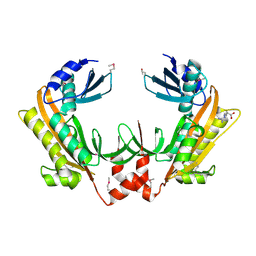 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ | | 分子名称: | ARGININE, GP37, POTASSIUM ION | | 著者 | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | 登録日 | 2016-01-08 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
6ZFP
 
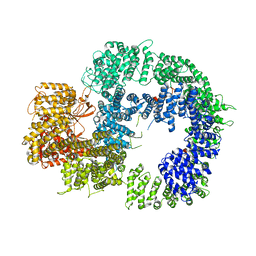 | | Cryo-EM structure of DNA-PKcs (State 2) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
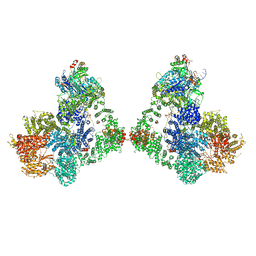 | | Cryo-EM structure of DNA-PK dimer | | 分子名称: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
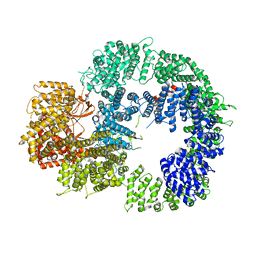 | | Cryo-EM structure of DNA-PKcs (State 1) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
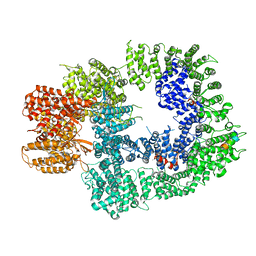 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
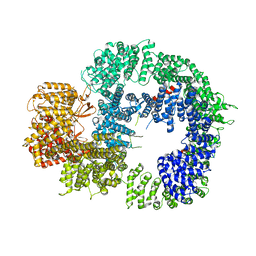 | | Cryo-EM structure of DNA-PKcs (State 3) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2YJV
 
 | |
6ZHA
 
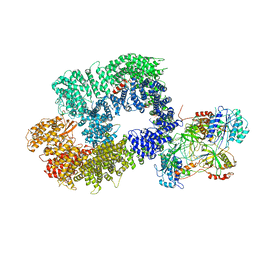 | | Cryo-EM structure of DNA-PK monomer | | 分子名称: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2YJT
 
 | |
6ZH8
 
 | | Cryo-EM structure of DNA-PKcs:DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
