1T92
 
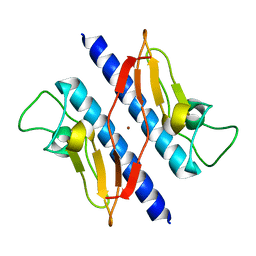 | | Crystal structure of N-terminal truncated pseudopilin PulG | | 分子名称: | General secretion pathway protein G, ZINC ION | | 著者 | Koehler, R, Schaefer, K, Mueller, S, Vignon, G, Diederichs, K, Philippsen, A, Ringler, P, Pugsley, A.P, Engel, A, Welte, W. | | 登録日 | 2004-05-14 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and assembly of the pseudopilin PulG.
Mol.Microbiol., 54, 2004
|
|
4OJ2
 
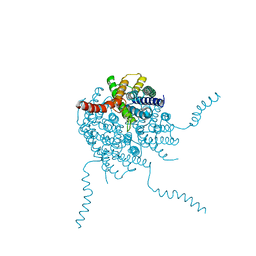 | |
1U0D
 
 | | Y33H Mutant of Homing endonuclease I-CreI | | 分子名称: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', DNA endonuclease I-CreI | | 著者 | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
7JTI
 
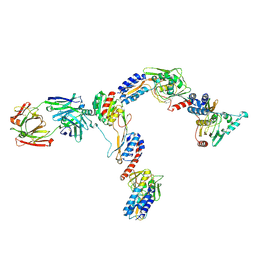 | | Interphotoreceptor retinoid-binding protein (IRBP) in complex with a monoclonal antibody (F3F5 mAb5) | | 分子名称: | Retinol-binding protein 3, mAb5 Fab heavy chain, mAb5 Fab light chain | | 著者 | Sears, A.E, Albiez, S, Gulati, S, Wang, B, Kiser, P, Kovacik, L, Engel, A, Stahlberg, H, Palczewski, K. | | 登録日 | 2020-08-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody.
Faseb J., 34, 2020
|
|
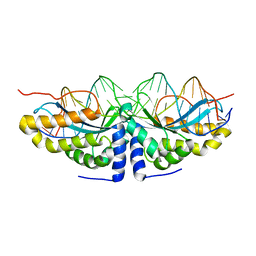
1U0C
 
 | | Y33C Mutant of Homing endonuclease I-CreI | | 分子名称: | 5'-D(*CP*GP*TP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*AP*CP*G)-3', DNA endonuclease I-CreI, ... | | 著者 | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
1H6I
 
 | |
1EIK
 
 | | Solution Structure of RNA Polymerase Subunit RPB5 from Methanobacterium Thermoautotrophicum | | 分子名称: | RNA POLYMERASE SUBUNIT RPB5 | | 著者 | Yee, A, Booth, V, Dharamsi, A, Engel, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-02-25 | | 公開日 | 2000-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RNA polymerase subunit RPB5 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FQY
 
 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | 分子名称: | AQUAPORIN-1 | | 著者 | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | 主引用文献 | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
6F7B
 
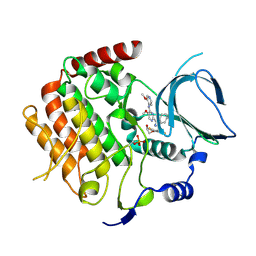 | | Crystal structure of the human Bub1 kinase domain in complex with BAY 1816032 | | 分子名称: | 2-[3,5-bis(fluoranyl)-4-[[3-[5-methoxy-4-[(3-methoxypyridin-4-yl)amino]pyrimidin-2-yl]indazol-1-yl]methyl]phenoxy]ethanol, MAGNESIUM ION, Mitotic checkpoint serine/threonine-protein kinase BUB1 | | 著者 | Holton, S.J, Siemeister, G, Mengel, A, Bone, W, Schroeder, J, Zitzmann-Kolbe, S, Briem, H, Fernandez-Montalvan, A, Prechtl, S, Moenning, U, von Ahsen, O, Johanssen, J, Cleve, A, Puetter, V, Hitchcock, M, von Nussbaum, F, Brands, M, Mumberg, D, Ziegelbauer, K. | | 登録日 | 2017-12-08 | | 公開日 | 2018-12-19 | | 最終更新日 | 2021-05-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP InhibitorsIn VitroandIn Vivo.
Clin.Cancer Res., 25, 2019
|
|
2JBP
 
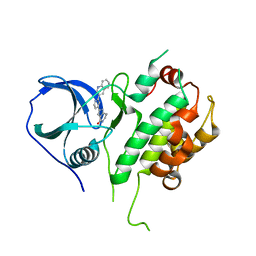 | | Protein kinase MK2 in complex with an inhibitor (crystal form-2, co- crystallization) | | 分子名称: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2 | | 著者 | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | 登録日 | 2006-12-09 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Structural basis for a high affinity inhibitor bound to protein kinase MK2.
J. Mol. Biol., 369, 2007
|
|
2JBO
 
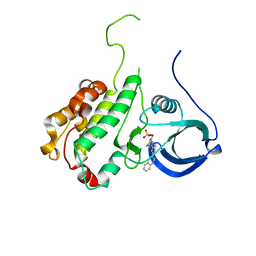 | | Protein kinase MK2 in complex with an inhibitor (crystal form-1, soaking) | | 分子名称: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2, PHOSPHATE ION | | 著者 | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | 登録日 | 2006-12-09 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for a High Affinity Inhibitor Bound to Protein Kinase Mk2.
J.Mol.Biol., 369, 2007
|
|
1Z9E
 
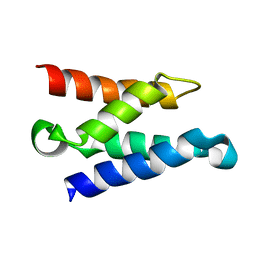 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | 分子名称: | PC4 and SFRS1 interacting protein 2 | | 著者 | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | 登録日 | 2005-04-01 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | 分子名称: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | 著者 | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | 登録日 | 2005-09-24 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ITG
 
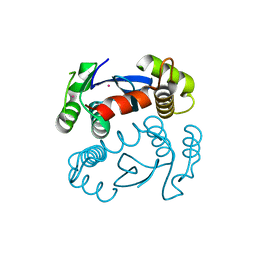 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | 分子名称: | CACODYLATE ION, HIV-1 INTEGRASE | | 著者 | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | 登録日 | 1994-11-21 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
6N1G
 
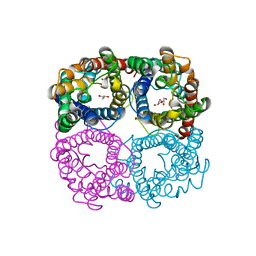 | |
6MZB
 
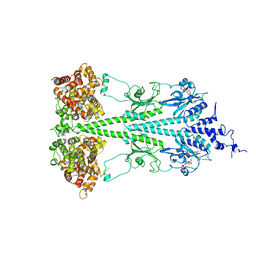 | | Cryo-EM structure of phosphodiesterase 6 | | 分子名称: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | 著者 | Gulati, S, Palczewski, K. | | 登録日 | 2018-11-04 | | 公開日 | 2019-03-06 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.
Sci Adv, 5, 2019
|
|
2RKO
 
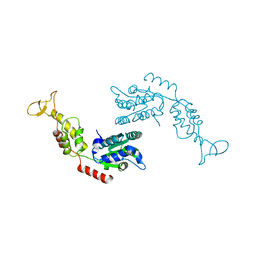 | |
7JTZ
 
 | | Yeast Glo3 GAP domain | | 分子名称: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | 著者 | Xie, B, Jackson, L.P. | | 登録日 | 2020-08-18 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
2C32
 
 | |
1GH8
 
 | |
5D7U
 
 | | Crystal structure of the C-terminal domain of MMTV integrase | | 分子名称: | ISOPROPYL ALCOHOL, Pr160 | | 著者 | Cook, N.J, Pye, V.E, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | 登録日 | 2015-08-14 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
4ID1
 
 | |
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | 分子名称: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | 著者 | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | 登録日 | 2015-07-31 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
4TSX
 
 | |
5CZ1
 
 | |
