8E1A
 
 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | 著者 | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | 登録日 | 2022-08-10 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
4HLW
 
 | | Targeting the Binding Function 3 (BF3) Site of the Human Androgen Receptor Through Virtual Screening. 2. Development of 2-((2-phenoxyethyl) thio)-1H-benzoimidazole derivatives. | | 分子名称: | 2-[(2-phenoxyethyl)sulfanyl]-1H-benzimidazole, Androgen receptor, GLYCEROL, ... | | 著者 | Ravi, S.N.M, Leblanc, E, Axerio-Cilies, P, Labriere, C, Frewin, K, Hassona, M.D.H, Lack, N.A, Han, F.Q, Guns, E.S, Young, R, Ban, F, Rennie, P.S, Cherkasov, A. | | 登録日 | 2012-10-17 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Targeting the Binding Function 3 (BF3) Site of the Androgen Receptor Through Virtual Screening. 2. Development of 2-((2-phenoxyethyl) thio)-1H-benzimidazole Derivatives.
J.Med.Chem., 56, 2013
|
|
2XM9
 
 | | Structure of a small molecule inhibitor with the kinase domain of Chk2 | | 分子名称: | 4-(1H-pyrazol-5-yl)-2-{4-[(3S)-pyrrolidin-3-ylamino]quinazolin-2-yl}phenol, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | 著者 | Caldwell, J.J, Welsh, E.J, Matijssen, C, Anderson, V.E, Antoni, L, Boxall, K, Urban, F, Hayes, A, Raynaud, F.I, Rigoreau, L.J, Raynham, T, Aherne, G.W, Pearl, L.H, Oliver, A.W, Garrett, M.D, Collins, I. | | 登録日 | 2010-07-26 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
2XM8
 
 | | Co-crystal structure of a small molecule inhibitor bound to the kinase domain of Chk2 | | 分子名称: | 2-{4-[(3S)-PYRROLIDIN-3-YLAMINO]QUINAZOLIN-2-YL}PHENOL, SERINE/THREONINE-PROTEIN KINASE CHK2 | | 著者 | Caldwell, J.J, Welsh, E.J, Matijssen, C, Anderson, V.E, Antoni, L, Boxall, K, Urban, F, Hayes, A, Raynaud, F.I, Rigoreau, L.J, Raynham, T, Aherne, G.W, Pearl, L.H, Oliver, A.W, Garrett, M.D, Collins, I. | | 登録日 | 2010-07-26 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
1PV2
 
 | |
1N57
 
 | | Crystal Structure of Chaperone Hsp31 | | 分子名称: | Chaperone Hsp31, MAGNESIUM ION | | 著者 | Quigley, P.M, Korotkov, K, Baneyx, F, Hol, W.G.J. | | 登録日 | 2002-11-04 | | 公開日 | 2003-03-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The 1.6A Crystal Structure of the Class of Chaperone Represented by
Escherichia coli Hsp31 Reveals a Putative Catalytic Triad
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2BJH
 
 | | Crystal Structure Of S133A AnFaeA-ferulic acid complex | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | 著者 | Faulds, C.B, Molina, R, Gonzalez, R, Husband, F, Juge, N, Sanz-Aparicio, J, Hermoso, J.A. | | 登録日 | 2005-02-02 | | 公開日 | 2005-09-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Probing the Determinants of Substrate Specificity of a Feruloyl Esterase, Anfaea, from Aspergillus Niger
FEBS J., 272, 2005
|
|
6DLG
 
 | | Crystal structure of a SHIP1 surface entropy reduction mutant | | 分子名称: | ISOPROPYL ALCOHOL, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Gardill, B.R, Cheung, S.T, Mui, A.L, Van Petegem, F. | | 登録日 | 2018-06-01 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Interleukin-10 and Small Molecule SHIP1 Allosteric Regulators Trigger Anti-Inflammatory Effects Through SHIP1/STAT3 Complexes
Biorxiv, 2020
|
|
6DV5
 
 | |
8SXR
 
 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | 分子名称: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | 登録日 | 2023-05-23 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
7JP1
 
 | |
7JOY
 
 | |
7KHP
 
 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2020-10-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
8CZ7
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYZ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYU
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | 分子名称: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | 分子名称: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | 登録日 | 2022-05-24 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRP
 
 | | Structure of the AaLpxC/LPC-023 Complex | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | 著者 | Najeeb, J, Lee, C.-J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
8E4A
 
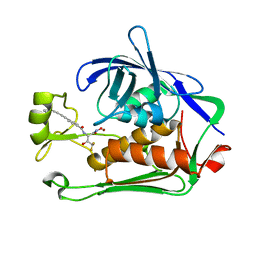 | | Pseudomonas LpxC in complex with LPC-233 | | 分子名称: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | 著者 | Najeeb, J, Zhou, P. | | 登録日 | 2022-08-17 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.034 Å) | | 主引用文献 | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
2B5Z
 
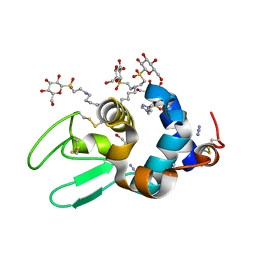 | | Hen lysozyme chemically glycosylated | | 分子名称: | (1S)-1,5-anhydro-1-(ethylsulfonyl)-D-glucitol, AZIDE ION, GLYCEROL, ... | | 著者 | Lopez-Jaramillo, F.J, Perez-Balderas, F, Hernandez-Mateo, F, Santoyo-Gonzalez, F. | | 登録日 | 2005-09-29 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Production, crystallization and X-ray characterization of chemically glycosylated hen egg-white lysozyme.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7U0O
 
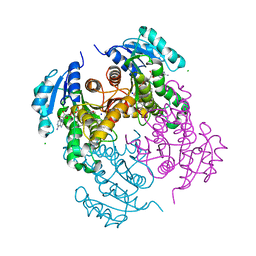 | |
5I8O
 
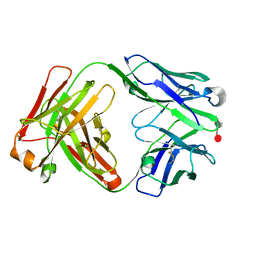 | |
7U0M
 
 | |
