5JPX
 
 | |
6VRG
 
 | | Structure of HIV-1 integrase with native amino-terminal sequence | | 分子名称: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | 登録日 | 2020-02-07 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
1NXM
 
 | | The high resolution structures of RmlC from Streptococcus suis | | 分子名称: | dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-11 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NYW
 
 | | The high resolution structures of RmlC from Streptoccus suis in complex with dTDP-D-glucose | | 分子名称: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-14 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NZC
 
 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | 分子名称: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-17 | | 公開日 | 2003-06-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
4GVB
 
 | |
6H1E
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH and H4 peptide | | 分子名称: | HemK methyltransferase family member 2, Histone H4 peptide, Multifunctional methyltransferase subunit TRM112-like protein, ... | | 著者 | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | 登録日 | 2018-07-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6H1D
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH | | 分子名称: | HemK methyltransferase family member 2, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | 登録日 | 2018-07-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7KQV
 
 | |
7KRG
 
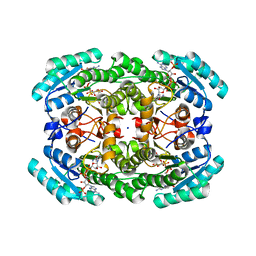 | |
6G9P
 
 | |
6E1J
 
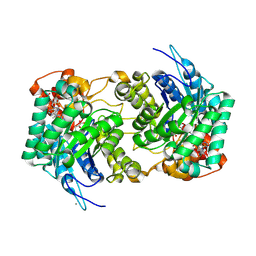 | |
8CT5
 
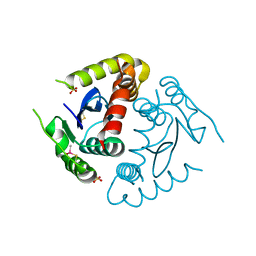 | |
8CTA
 
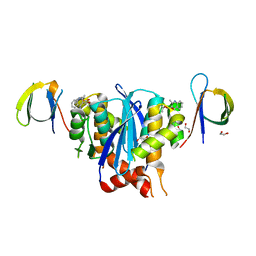 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | 分子名称: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | 著者 | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8CT7
 
 | | Catalytic Core Domain of HIV-1 Integrase (F185K) bound with BI-224436 | | 分子名称: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | 著者 | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
6FGA
 
 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | 分子名称: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | 著者 | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | 登録日 | 2018-01-10 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | 分子名称: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | 登録日 | 2021-09-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUL
 
 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | 分子名称: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | 著者 | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | 登録日 | 2021-09-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | 分子名称: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | 著者 | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | 登録日 | 2021-09-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
6O8I
 
 | | BTK In Complex With Inhibitor | | 分子名称: | 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, Tyrosine-protein kinase BTK | | 著者 | Pokross, M, Tebben, A.J, Watterson, S.H. | | 登録日 | 2019-03-11 | | 公開日 | 2019-04-03 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of Branebrutinib (BMS-986195): A Strategy for Identifying a Highly Potent and Selective Covalent Inhibitor Providing Rapid in Vivo Inactivation of Bruton's Tyrosine Kinase (BTK).
J. Med. Chem., 62, 2019
|
|
6AZW
 
 | | IDO1/FXB-001116 crystal structure | | 分子名称: | (2R)-N-(4-cyanophenyl)-2-[cis-4-(quinolin-4-yl)cyclohexyl]propanamide, Indoleamine 2,3-dioxygenase 1 | | 著者 | Lewis, H.A, Lammens, A, Steinbacher, S. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AZV
 
 | | IDO1/BMS-978587 crystal structure | | 分子名称: | (1R,2S)-2-(4-[bis(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, Indoleamine 2,3-dioxygenase 1 | | 著者 | Lewis, H.A. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.755 Å) | | 主引用文献 | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AZU
 
 | | Holo IDO1 crystal structure | | 分子名称: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Lewis, H.A, Yan, C. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.822 Å) | | 主引用文献 | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E24
 
 | | Ternary structure of c-Myc-TBP-TAF1 | | 分子名称: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | 著者 | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2018-07-10 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6E16
 
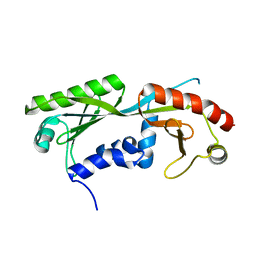 | | Ternary structure of c-Myc-TBP-TAF1 | | 分子名称: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | 著者 | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2018-07-09 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
