7VNH
 
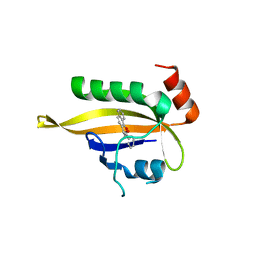 | |
7VRE
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | 分子名称: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | 著者 | Zhu, S.J. | | 登録日 | 2021-10-22 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.507 Å) | | 主引用文献 | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
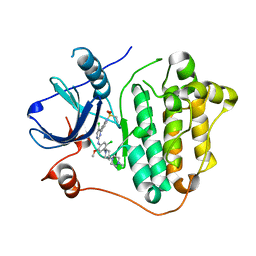
7VRA
 
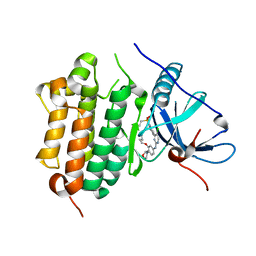 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | 分子名称: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | 著者 | Zhu, S.J. | | 登録日 | 2021-10-22 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7V29
 
 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | 分子名称: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | 著者 | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | 登録日 | 2021-08-07 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | 分子名称: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | 著者 | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | 登録日 | 2021-02-21 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
3SJQ
 
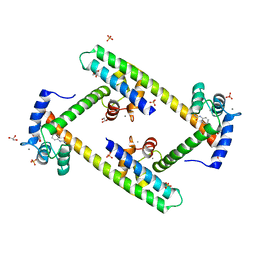 | | Crystal structure of a small conductance potassium channel splice variant complexed with calcium-calmodulin | | 分子名称: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | 著者 | Zhang, M, Pascal, J.M, Zhang, J.-F. | | 登録日 | 2011-06-21 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for calmodulin as a dynamic calcium sensor.
Structure, 20, 2012
|
|
5VZ4
 
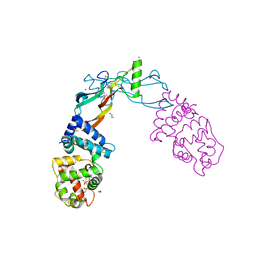 | | Receptor-growth factor crystal structure at 2.20 Angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, GDNF family receptor alpha-like, ... | | 著者 | Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2017-05-26 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15.
Nature, 550, 2017
|
|
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Wang, X, Cao, L. | | 登録日 | 2021-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
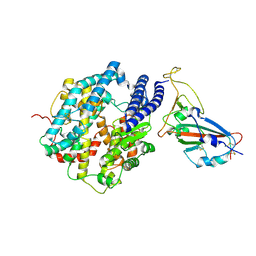 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Wang, X, Cao, L. | | 登録日 | 2021-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
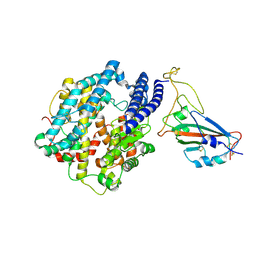 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Wang, X, Cao, L. | | 登録日 | 2021-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDG
 
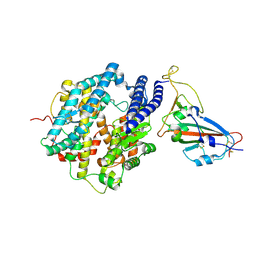 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Wang, X, Cao, L. | | 登録日 | 2021-07-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7C43
 
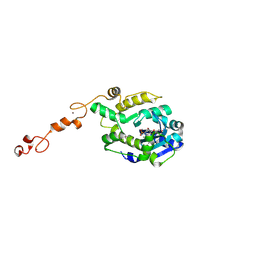 | |
7C4C
 
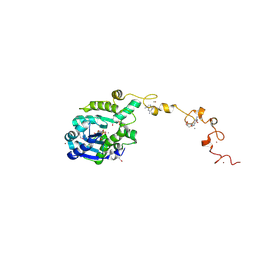 | |
7C4B
 
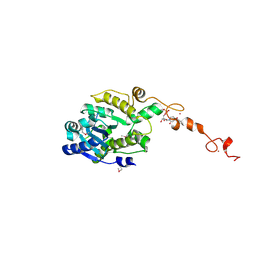 | |
7C45
 
 | |
7C47
 
 | |
7C42
 
 | |
3RBQ
 
 | | Co-crystal structure of human UNC119 (retina gene 4) and an N-terminal Transducin-alpha mimicking peptide | | 分子名称: | Guanine nucleotide-binding protein G(t) subunit alpha-1, Protein unc-119 homolog A | | 著者 | Constantine, R, Whitby, F.G, Hill, C.P, Baehr, W. | | 登録日 | 2011-03-29 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | UNC119 is required for G protein trafficking in sensory neurons.
Nat.Neurosci., 14, 2011
|
|
7WZX
 
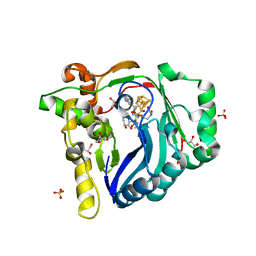 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.980013 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
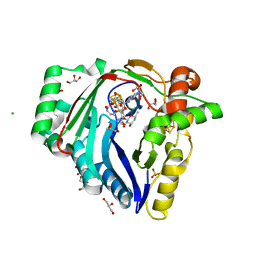 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | 著者 | Zhou, J.H, Hou, X.L. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.899313 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
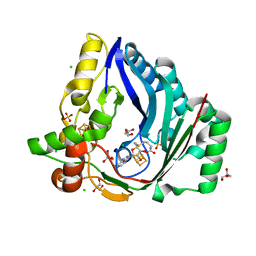 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02027535 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
4ANE
 
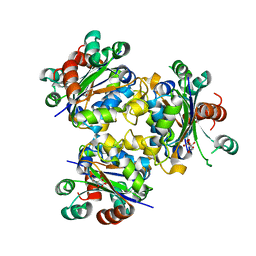 | | R80N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | CITRIC ACID, NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | 登録日 | 2012-03-16 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Mycobacterium Tuberculosis Nucleoside Diphosphate Kinase R80N Mutant in Complex with Citrate
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ANC
 
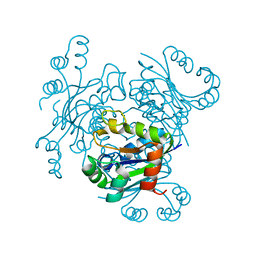 | | CRYSTAL FORM I OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | 登録日 | 2012-03-16 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
4AND
 
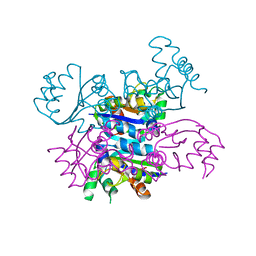 | | CRYSTAL FORM II OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | 登録日 | 2012-03-16 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.808 Å) | | 主引用文献 | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
3RPM
 
 | |
