6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | 分子名称: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2019-03-30 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | 分子名称: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | 登録日 | 2019-05-07 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
5I3P
 
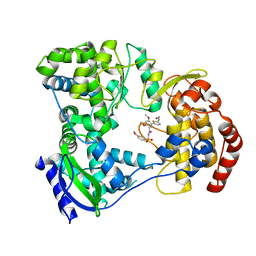 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 27 | | 分子名称: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-2,4-dimethoxy-N-[(3-methoxyphenyl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | 著者 | Noble, C.G. | | 登録日 | 2016-02-10 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5I3Q
 
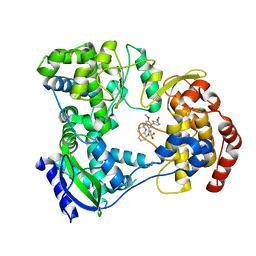 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 29 | | 分子名称: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | 著者 | Noble, C.G. | | 登録日 | 2016-02-10 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
7WPV
 
 | |
7WPH
 
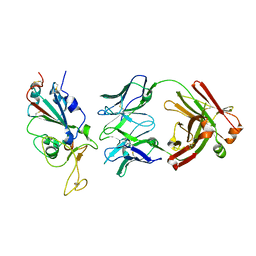 | | SARS-CoV2 RBD bound to Fab06 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | 著者 | Lin, J.Q, El Sahili, A, Lescar, J. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
8GB0
 
 | |
8JPX
 
 | |
5Z7L
 
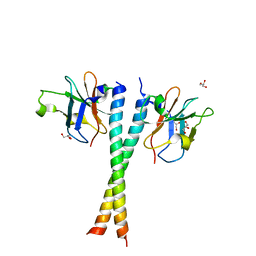 | | Crystal structure of NDP52 SKICH region in complex with NAP1 | | 分子名称: | 5-azacytidine-induced protein 2, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL | | 著者 | Fu, T, Pan, L.F. | | 登録日 | 2018-01-29 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YUP
 
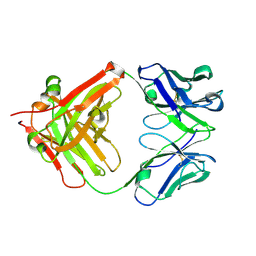 | | Crystal Structure of the Fab fragment of FVIIa antibody mAb4F5 | | 分子名称: | the heavy chain of the Fab fragment of FVIIa antibody mAb4F5, the light chain of the Fab fragment of FVIIa antibody mAb4F5 | | 著者 | Jiang, L.G, Persson, E, Huang, M.D. | | 登録日 | 2017-11-22 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure, epitope, and functional impact of an antibody against a superactive FVIIa provide insights into allosteric mechanism.
Res Pract Thromb Haemost, 3, 2019
|
|
6A7U
 
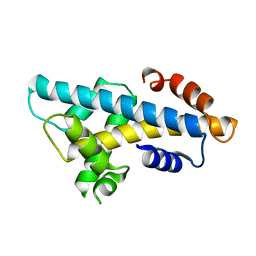 | | Crystal structure of histone H2A.Bbd-H2B dimer | | 分子名称: | Histone H2B type 2-E,Histone H2A-Bbd type 2/3 | | 著者 | Dai, L, Zhou, Z. | | 登録日 | 2018-07-04 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the histone heterodimer containing histone variant H2A.Bbd.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
8K67
 
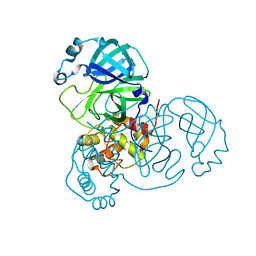 | |
8K6B
 
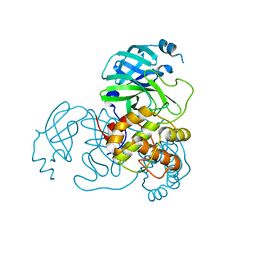 | |
8K6A
 
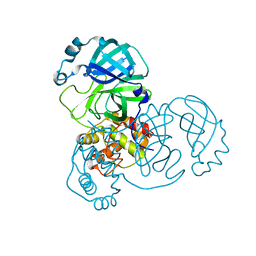 | |
8K6C
 
 | |
8K68
 
 | |
8K6D
 
 | |
6AKY
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | 著者 | Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2018-09-04 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
6AKX
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 | | 分子名称: | C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-2-yl)propyl]cyclopentanecarboxamide, NITRATE ION, ... | | 著者 | Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2018-09-04 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
2JC2
 
 | | The crystal structure of the natural F112L human sorcin mutant | | 分子名称: | SORCIN, SULFATE ION | | 著者 | Franceschini, S, Ilari, A, Colotti, G, Chiancone, E. | | 登録日 | 2006-12-19 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular Basis for the Impaired Function of the Natural F112L Sorcin Mutant: X-Ray Crystal Structure, Calcium Affinity, and Interaction with Annexin Vii and the Ryanodine Receptor.
Faseb J., 22, 2008
|
|
6IUH
 
 | |
6IUI
 
 | |
7XXL
 
 | | RBD in complex with Fab14 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | 著者 | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | 登録日 | 2022-05-30 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7F2M
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18d | | 分子名称: | (~{Z})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | 著者 | Huang, Y.-Y, He, X, Luo, H.-B. | | 登録日 | 2021-06-11 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.20004153 Å) | | 主引用文献 | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
