3PFQ
 
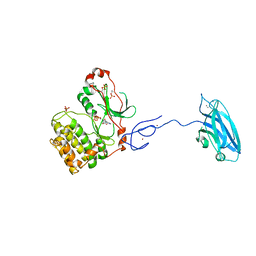 | | Crystal Structure and Allosteric Activation of Protein Kinase C beta II | | 分子名称: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase C beta type, ... | | 著者 | Leonard, T.A, Rozycki, B, Saidi, L.F, Hummer, G, Hurley, J.H. | | 登録日 | 2010-10-28 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Cell(Cambridge,Mass.), 144, 2011
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | 分子名称: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | 著者 | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | 登録日 | 2022-02-10 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | 分子名称: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | 著者 | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | 登録日 | 2022-02-10 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (50 Å) | | 主引用文献 | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
3F1I
 
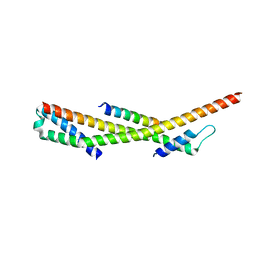 | | Human ESCRT-0 Core Complex | | 分子名称: | Hepatocyte growth factor-regulated tyrosine kinase substrate, Signal transducing adapter molecule 1 | | 著者 | Ren, X, Kloer, D.P, Kim, Y, Ghirlando, R, Saidi, L, Hummer, G, Hurley, J.H. | | 登録日 | 2008-10-28 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Hybrid Structural Model of the Complete Human ESCRT-0 Complex.
Structure, 17, 2009
|
|
1M6I
 
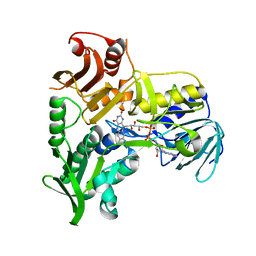 | | Crystal Structure of Apoptosis Inducing Factor (AIF) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Programmed cell death protein 8 | | 著者 | Ye, H, Cande, C, Stephanou, N.C, Jiang, S, Gurbuxani, S, Larochette, N, Daugas, E, Garrido, C, Kroemer, G, Wu, H. | | 登録日 | 2002-07-16 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | DNA binding is required for the apoptogenic action of apoptosis inducing factor.
Nat.Struct.Biol., 9, 2002
|
|
4JVB
 
 | | Crystal structure of PDE6D in complex with the inhibitor rac-2 | | 分子名称: | 1-benzyl-2-(4-{[(2R)-2-(2-phenyl-1H-benzimidazol-1-yl)pent-4-en-1-yl]oxy}phenyl)-1H-benzimidazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | 著者 | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
4JVF
 
 | | The Crystal structure of PDE6D in complex with the inhibitor (s)-5 | | 分子名称: | (2S)-2-(2-phenyl-1H-benzimidazol-1-yl)-2-(piperidin-4-yl)ethyl 1-(1-benzyl-1H-benzimidazol-2-yl)piperidine-4-carboxylate, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | 著者 | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
4JV6
 
 | | The crystal structure of PDE6D in complex to inhibitor-1 | | 分子名称: | 1-benzyl-2-phenyl-1H-benzimidazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | 著者 | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
4JV8
 
 | | The crystal structure of PDE6D in complex with rac-S1 | | 分子名称: | (6R)-6-(pyridin-2-yl)-5,6-dihydrobenzimidazo[1,2-c]quinazoline, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | 著者 | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
2VU1
 
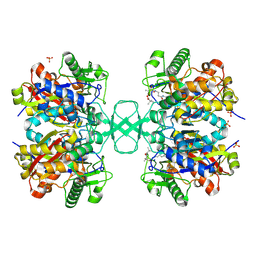 | | Biosynthetic thiolase from Z. ramigera. Complex of with O-pantheteine- 11-pivalate. | | 分子名称: | ACETYL-COA ACETYLTRANSFERASE, PANTOTHENYL-AMINOETHANOL-11-PIVALIC ACID, SODIUM ION, ... | | 著者 | Kursula, P, Schmitz, W, Wierenga, R.K. | | 登録日 | 2008-05-19 | | 公開日 | 2008-10-28 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
2VU0
 
 | |
8S9K
 
 | | Structure of dimeric FAM111A SPD S541A Mutant | | 分子名称: | GLYCEROL, Serine protease FAM111A | | 著者 | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | 登録日 | 2023-03-29 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
8S9L
 
 | | Structure of monomeric FAM111A SPD V347D Mutant | | 分子名称: | SULFATE ION, Serine protease FAM111A | | 著者 | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | 登録日 | 2023-03-29 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
6YVA
 
 | | PLpro-C111S with mISG15 | | 分子名称: | Replicase polyprotein 1a, Ubiquitin-like protein ISG15, ZINC ION | | 著者 | Shin, D, Dikic, I. | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity.
Nature, 587, 2020
|
|
9RUB
 
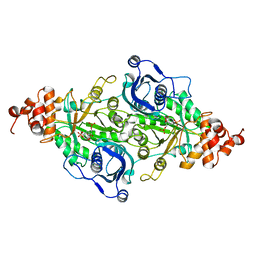 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | 分子名称: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | 著者 | Lundqvist, T, Schneider, G. | | 登録日 | 1990-11-28 | | 公開日 | 1993-01-15 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
1VRH
 
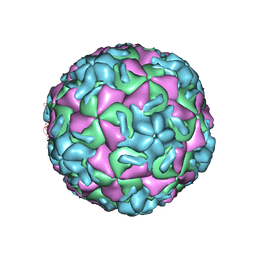 | | HRV14/SDZ 880-061 COMPLEX | | 分子名称: | 2-[4-(2H-1,4-BENZOTHIAZINE-3-YL)-PIPERAZINE-1-LY]-1,3-THIAZOLE-4-CARBOXYLIC ACID ETHYLESTER, RHINOVIRUS 14 | | 著者 | Oren, D.A, Zhang, A, Arnold, E. | | 登録日 | 1996-02-26 | | 公開日 | 1997-02-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Synthesis and activity of piperazine-containing antirhinoviral agents and crystal structure of SDZ 880-061 bound to human rhinovirus 14.
J.Mol.Biol., 259, 1996
|
|
1VSM
 
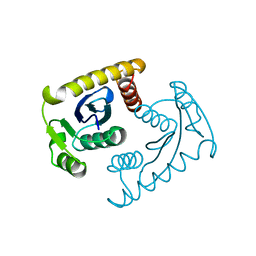 | |
1VSK
 
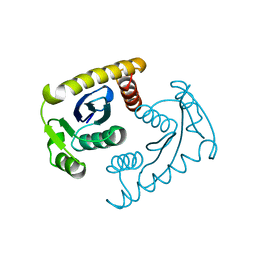 | |
1MEC
 
 | |
7LP4
 
 | | Structure of Nedd4L WW3 domain | | 分子名称: | E3 ubiquitin-protein ligase NEDD4-like | | 著者 | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | 登録日 | 2021-02-11 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
5D5R
 
 | |
8R8T
 
 | |
3H9E
 
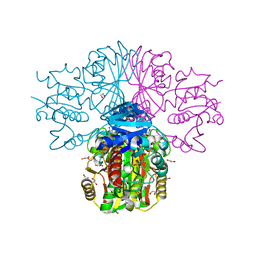 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | 分子名称: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | 著者 | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2009-04-30 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
4F0Z
 
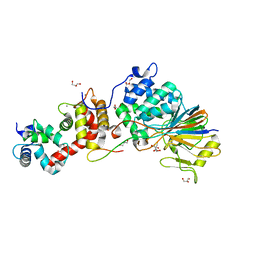 | | Crystal Structure of Calcineurin in Complex with the Calcineurin-Inhibiting Domain of the African Swine Fever Virus Protein A238L | | 分子名称: | Ankyrin repeat domain-containing protein A238L, CALCIUM ION, Calcineurin subunit B type 1, ... | | 著者 | Grigoriu, S, Peti, W, Page, R. | | 登録日 | 2012-05-05 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin.
Plos Biol., 11, 2013
|
|
4X34
 
 | |
