3HMQ
 
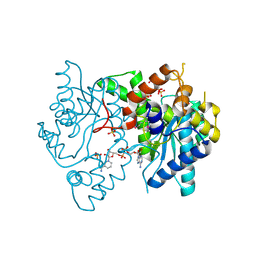 | | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+) | | 分子名称: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-05-29 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+)
To be Published
|
|
6MXV
 
 | | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | 著者 | Tan, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-31 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
6NFP
 
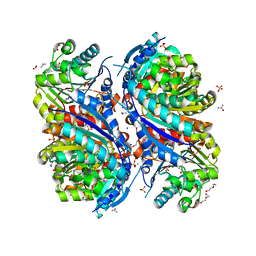 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | 分子名称: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | 著者 | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-12-20 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6OVW
 
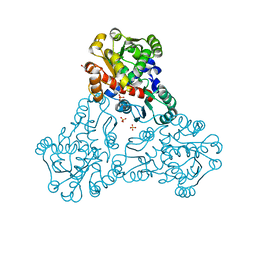 | | Crystal structure of ornithine carbamoyltransferase from Salmonella enterica | | 分子名称: | GLYCEROL, Ornithine carbamoyltransferase, PHOSPHATE ION | | 著者 | Chang, C, Mesa, N, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-08 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Crystal structure of ornithine carbamoyltransferase from Salmonella enterica
To Be Published
|
|
2G8Y
 
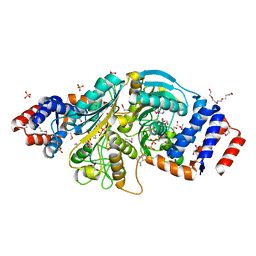 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | 分子名称: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-03-03 | | 公開日 | 2006-04-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
1X7V
 
 | | Crystal structure of PA3566 from Pseudomonas aeruginosa | | 分子名称: | PA3566 protein, SULFATE ION | | 著者 | Sanders, D.A, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-08-16 | | 公開日 | 2004-08-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The X-ray crystal structure of PA3566 from Pseudomonas aureginosa at 1.8 A resolution.
Proteins, 61, 2005
|
|
6AOK
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | 著者 | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | 登録日 | 2017-08-16 | | 公開日 | 2018-01-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
1XSV
 
 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | 分子名称: | Hypothetical UPF0122 protein SAV1236 | | 著者 | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | 登録日 | 2004-10-20 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
1TE2
 
 | | Putative Phosphatase Ynic from Escherichia coli K12 | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, 2-deoxyglucose-6-P phosphatase, CALCIUM ION | | 著者 | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-05-24 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal Structure of Putative Phosphatase Ynic from Escherichia coli K12
To be Published
|
|
4U12
 
 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | 分子名称: | Uncharacterized protein HP0242 | | 著者 | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-07-14 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
6P2K
 
 | | Crystal structure of AFV00434, an ancestral GH74 enzyme | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
4Q3K
 
 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6P2O
 
 | | Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
2KKY
 
 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | 分子名称: | Uncharacterized protein ECs2156 | | 著者 | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2009-06-29 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | GLYCEROL, MGS-M2 | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6P2L
 
 | | Crystal structure of Niastella koreensis GH74 (NkGH74) enzyme | | 分子名称: | CHLORIDE ION, Glycosyl hydrolase BNR repeat-containing protein, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
4Q3N
 
 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | 分子名称: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | 著者 | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-12-09 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
5T2X
 
 | | Crystal structure of Uncharacterised protein lpg1670 | | 分子名称: | Uncharacterized protein LPG1670 | | 著者 | Chang, C, Xu, X, Cui, H, SAVCHENKO, A, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-08-24 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Crystal structure of Uncharacterised protein lpg1670
To Be Published
|
|
2FTP
 
 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | 分子名称: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | 著者 | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-01-24 | | 公開日 | 2006-03-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
4Q3M
 
 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | MGS-M4, SODIUM ION, SULFATE ION | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
2KKX
 
 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | 分子名称: | Uncharacterized protein ECs2156 | | 著者 | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-06-29 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-21 | | 公開日 | 2014-05-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
6MN5
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | 分子名称: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
