8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
7MLZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | 分子名称: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | 著者 | Zhou, T, Tsybovsky, T, Kwong, P.D. | | 登録日 | 2021-04-29 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
8KAE
 
 | | 16d-bound human SPNS2 | | 分子名称: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | 著者 | He, Y, Duan, Y. | | 登録日 | 2023-08-03 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
6NZH
 
 | |
6NZF
 
 | |
6NZE
 
 | |
6OIE
 
 | |
6ZOZ
 
 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3W0K
 
 | |
7AMT
 
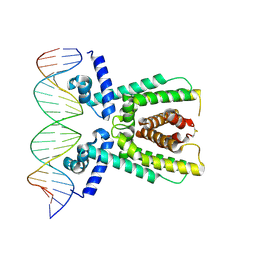 | | Structure of LuxR with DNA (activation) | | 分子名称: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | 著者 | Liu, B, Reverter, D. | | 登録日 | 2020-10-09 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7AMN
 
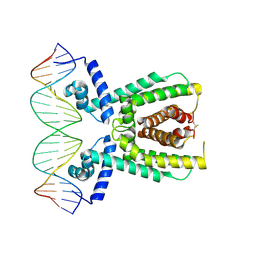 | | Structure of LuxR with DNA (repression) | | 分子名称: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | 著者 | Liu, B, Reverter, D. | | 登録日 | 2020-10-09 | | 公開日 | 2021-03-31 | | 最終更新日 | 2021-04-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
6ZP1
 
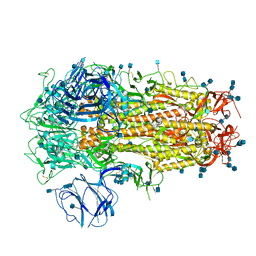 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QG1
 
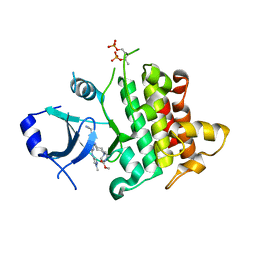 | | IRAK4 in complex with inhibitor | | 分子名称: | Interleukin-1 receptor-associated kinase 4, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]-5-oxidanylidene-pyrido[4,3-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | 著者 | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | 登録日 | 2021-12-07 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG3
 
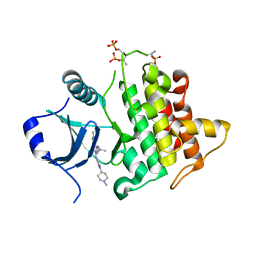 | | IRAK4 in complex with inhibitor | | 分子名称: | 6-[(2~{S})-2-fluoranylpropyl]-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | 著者 | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | 登録日 | 2021-12-07 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
6ZP0
 
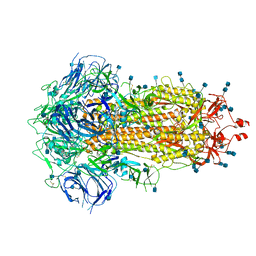 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOX
 
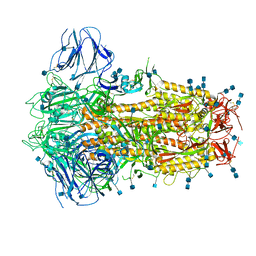 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
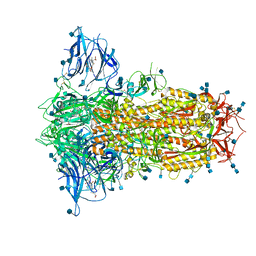 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOY
 
 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | 分子名称: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | 著者 | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | 登録日 | 2021-12-22 | | 公開日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | 分子名称: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | 著者 | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | 登録日 | 2021-12-21 | | 公開日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | 分子名称: | Frizzled-7,inhibitor peptide Fz7-21 | | 著者 | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | 登録日 | 2017-06-29 | | 公開日 | 2018-04-18 | | 最終更新日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
2JQB
 
 | | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5 | | 分子名称: | Conomarphin | | 著者 | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | 登録日 | 2007-05-31 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5
To be Published
|
|
7OKB
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 45 | | 分子名称: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, SULFATE ION, ... | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.58 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKC
 
 | | Crystal structure of Escherichia coli LpxA in complex with compound 1 | | 分子名称: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SODIUM ION | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKA
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 14 | | 分子名称: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-(1~{H}-imidazol-4-ylmethyl)ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ... | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
