7WED
 
 | |
7WEA
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | 著者 | Wang, X, Wang, L. | | 登録日 | 2021-12-23 | | 公開日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
3OKQ
 
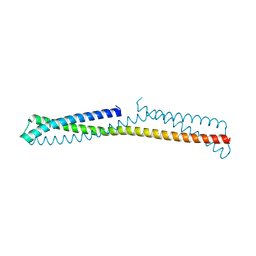 | |
3ONX
 
 | |
3TA3
 
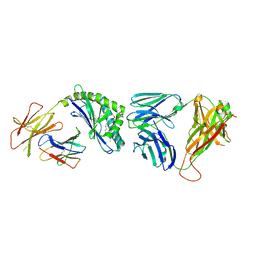 | | Structure of the mouse CD1d-Glc-DAG-s2-iNKT TCR complex | | 分子名称: | (2S)-1-(alpha-D-glucopyranosyloxy)-3-(hexadecanoyloxy)propan-2-yl (11Z)-octadec-11-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Girardi, E, Yu, E.D, Zajonc, D.M. | | 登録日 | 2011-08-03 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells.
Plos Biol., 9, 2011
|
|
3P3E
 
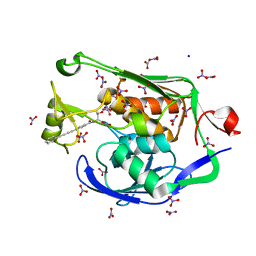 | | Crystal Structure of the PSEUDOMONAS AERUGINOSA LpxC/LPC-009 complex | | 分子名称: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, NITRATE ION, SODIUM ION, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-10-04 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
3P3G
 
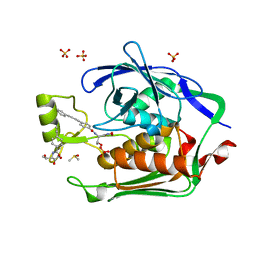 | | Crystal Structure of the Escherichia coli LpxC/LPC-009 complex | | 分子名称: | 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-10-04 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
3OQV
 
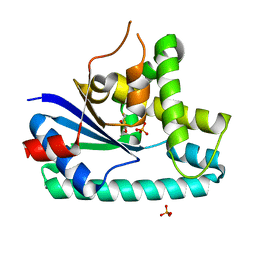 | | AlbC, a cyclodipeptide synthase from Streptomyces noursei | | 分子名称: | AlbC, DITHIANE DIOL, PHOSPHATE ION | | 著者 | Sauguet, L, Ledu, M.H, Charbonnier, J.B, Gondry, M. | | 登録日 | 2010-09-04 | | 公開日 | 2011-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis.
Nucleic Acids Res., 39, 2011
|
|
5X5O
 
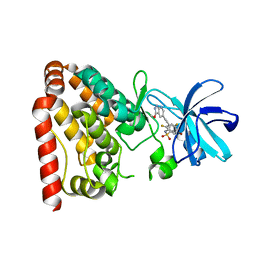 | | Crystal structure of ZAK in complex with compound D2829 | | 分子名称: | Mitogen-activated protein kinase kinase kinase MLT, N-[2,4-bis(fluoranyl)-3-[2-(3-methoxy-1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]phenyl]-3-bromanyl-benzenesulfonamide | | 著者 | Dai, Y.B, Zhao, P, Yun, C.H. | | 登録日 | 2017-02-17 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]pyridin-5-yl)ethynyl)benzenesulfonamides as Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J. Med. Chem., 60, 2017
|
|
3PS2
 
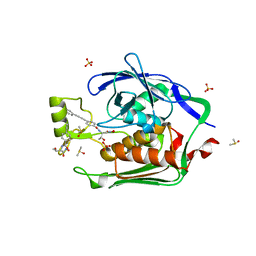 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | 分子名称: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | 著者 | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | 登録日 | 2020-09-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
5X5M
 
 | |
7X88
 
 | |
4RNG
 
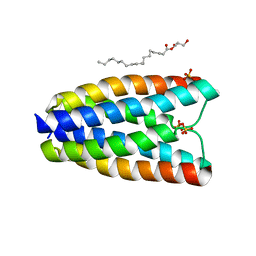 | | Crystal structure of a bacterial homologue of SWEET transporters | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MtN3/saliva family, SULFATE ION | | 著者 | Hu, Q, Wang, J, Yan, C, Yan, N. | | 登録日 | 2014-10-24 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of SWEET transporters.
Cell Res., 24, 2014
|
|
3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3OOI
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | 著者 | Qiao, Q, Wang, M, Xu, R.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | 著者 | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2022-11-17 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HHU
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with SY110 | | 分子名称: | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide, 3C-like proteinase nsp5 | | 著者 | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2022-11-17 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.258 Å) | | 主引用文献 | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
7K9X
 
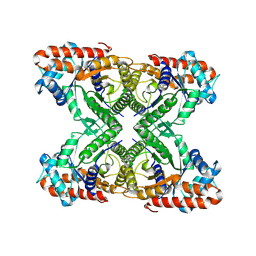 | |
8HN9
 
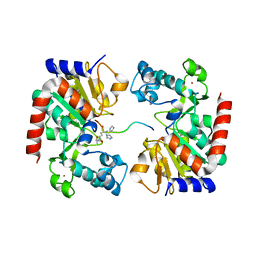 | | Human SIRT3 Recognizing CCNE2K348la peptide | | 分子名称: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | 著者 | Wang, Y, Ding, W. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
1BT7
 
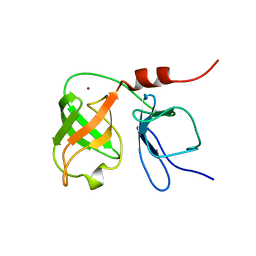 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | 分子名称: | NS3 SERINE PROTEASE, ZINC ION | | 著者 | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | 登録日 | 1998-09-01 | | 公開日 | 1999-06-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | 分子名称: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | 著者 | Zhou, X. | | 登録日 | 2022-07-01 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
8XKV
 
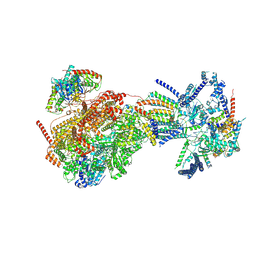 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in Apo state | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, chloroplastic, ... | | 著者 | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | 登録日 | 2023-12-25 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into the chloroplast protein import in land plants.
Cell, 2024
|
|
8XKU
 
 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in ATP-bound state | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, ... | | 著者 | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | 登録日 | 2023-12-24 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into the chloroplast protein import in land plants.
Cell, 2024
|
|
