6G2Y
 
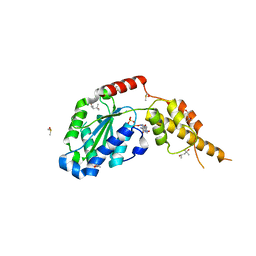 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2Z
 
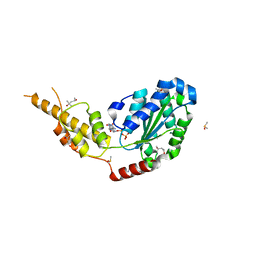 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
4XW2
 
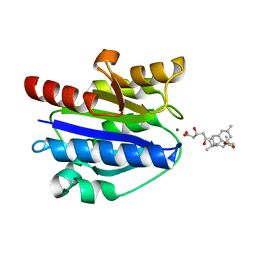 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | 分子名称: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | 著者 | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | 登録日 | 2015-01-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2014-04-09 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
4Z7P
 
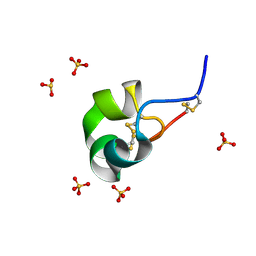 | | X-ray structure of racemic ShK Q16K toxin | | 分子名称: | Potassium channel toxin kappa-stichotoxin-She1a, SULFATE ION | | 著者 | Sickmier, E.A. | | 登録日 | 2015-04-07 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-09-23 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Pharmaceutical Optimization of Peptide Toxins for Ion Channel Targets: Potent, Selective, and Long-Lived Antagonists of Kv1.3.
J.Med.Chem., 58, 2015
|
|
5UND
 
 | |
1RUO
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | 著者 | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 1996-05-26 | | 公開日 | 1997-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
5V3G
 
 | | PRDM9-allele-C ZnF8-13 | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-03-07 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.416 Å) | | 主引用文献 | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
8GQU
 
 | | AK-42 inhibitor binding human ClC-2 TMD | | 分子名称: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | 著者 | Wang, L. | | 登録日 | 2022-08-30 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
7V6Y
 
 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | 分子名称: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
6IFN
 
 | | Crystal structure of Type III-A CRISPR Csm complex | | 分子名称: | MANGANESE (II) ION, RNA (32-MER), Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Wang, J, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
4X0N
 
 | |
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | 分子名称: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | 登録日 | 2011-11-04 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3JUA
 
 | |
6IZG
 
 | |
4EMM
 
 | |
1DDJ
 
 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | 分子名称: | PLASMINOGEN | | 著者 | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | 登録日 | 1999-11-10 | | 公開日 | 2000-02-18 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | 分子名称: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | 著者 | Chowdhury, R, Miller, M. | | 登録日 | 2021-12-20 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | 分子名称: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | 著者 | Chowdhury, R, Miller, M. | | 登録日 | 2021-12-20 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
5VNP
 
 | |
5LBV
 
 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | 分子名称: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | 著者 | Barba-Spaeth, G. | | 登録日 | 2016-06-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5L30
 
 | | Factor VIIa in complex with the inhibitor (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-4,15,17-trimethyl-7-[1-(1H-tetrazol-5-yl)cyclopropyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione | | 分子名称: | (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-4,15,17-trimethyl-7-[1-(1H-tetrazol-5-yl)cyclopropyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione, CALCIUM ION, Coagulation factor VII (Heavy Chain), ... | | 著者 | Wei, A. | | 登録日 | 2016-08-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8X3R
 
 | |
5LBS
 
 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | 分子名称: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | 著者 | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | 登録日 | 2016-06-17 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
