7TBI
 
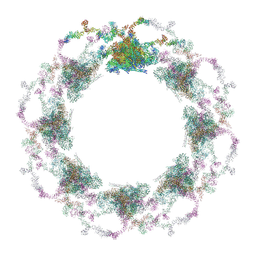 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
4XIC
 
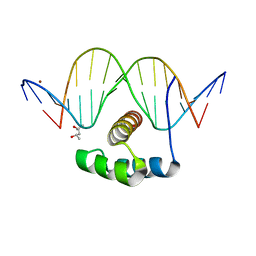 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | 著者 | White, M.A, Zandarashvili, L, Iwahara, J. | | 登録日 | 2015-01-06 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
4XW2
 
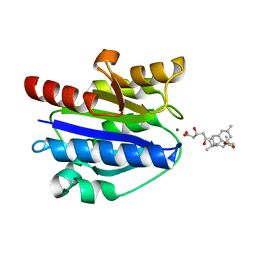 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | 分子名称: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | 著者 | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | 登録日 | 2015-01-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2014-04-09 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
5TJ7
 
 | |
5TJQ
 
 | | Structure of WWP2 2,3-linker-HECT | | 分子名称: | NEDD4-like E3 ubiquitin-protein ligase WWP2,NEDD4-like E3 ubiquitin-protein ligase WWP2 | | 著者 | Chen, Z, Gabelli, S.B. | | 登録日 | 2016-10-04 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A Tunable Brake for HECT Ubiquitin Ligases.
Mol. Cell, 66, 2017
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | 分子名称: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MNP
 
 | | Crystal structure of KPC-2 with compound 6 | | 分子名称: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2018-10-02 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5V3G
 
 | | PRDM9-allele-C ZnF8-13 | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-03-07 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.416 Å) | | 主引用文献 | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
6MLL
 
 | | Crystal structure of KPC-2 with compound 7 | | 分子名称: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2018-09-27 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | 分子名称: | Uncharacterized protein AF_1514 | | 著者 | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | 登録日 | 2008-01-20 | | 公開日 | 2008-02-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
5UY1
 
 | | X-ray crystal structure of apo Halotag | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Dunham, N.P, Boal, A.K. | | 登録日 | 2017-02-23 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
5TJ8
 
 | |
5UXZ
 
 | |
5UND
 
 | |
2LI6
 
 | |
1DDJ
 
 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | 分子名称: | PLASMINOGEN | | 著者 | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | 登録日 | 1999-11-10 | | 公開日 | 2000-02-18 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
5ZYA
 
 | | SF3b spliceosomal complex bound to E7107 | | 分子名称: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | 著者 | Finci, L.I, Larsen, N.A. | | 登録日 | 2018-05-23 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
6ACE
 
 | |
8KI4
 
 | | Crystal structure of human HSP90 in intermediate state | | 分子名称: | 1,2-ETHANEDIOL, Heat shock protein HSP 90-alpha | | 著者 | Xu, C, Zhang, X.L, Bai, F. | | 登録日 | 2023-08-22 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
5B6K
 
 | |
7DRT
 
 | | Human Wntless in complex with Wnt3a | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | 登録日 | 2020-12-29 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-09-08 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
